Slawomir Michniewski
@smichniewski
PhD | Bioinformatics | Phages | Vibrio | Klebsiella | AMR
ID: 971336609730834437
07-03-2018 10:47:49
85 Tweet
288 Followers
168 Following

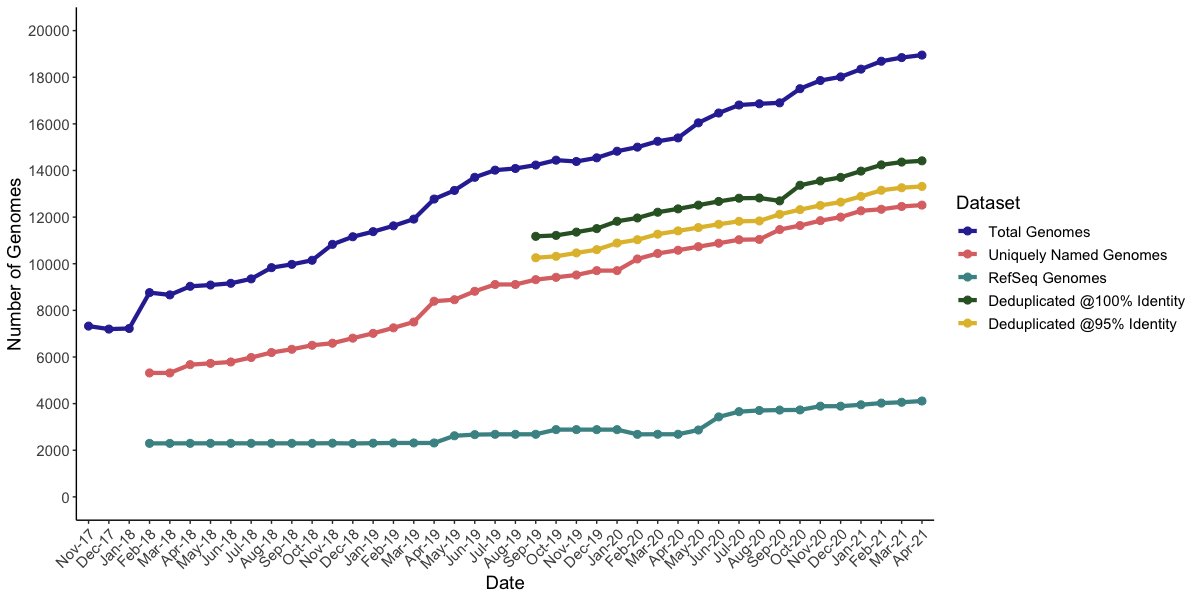
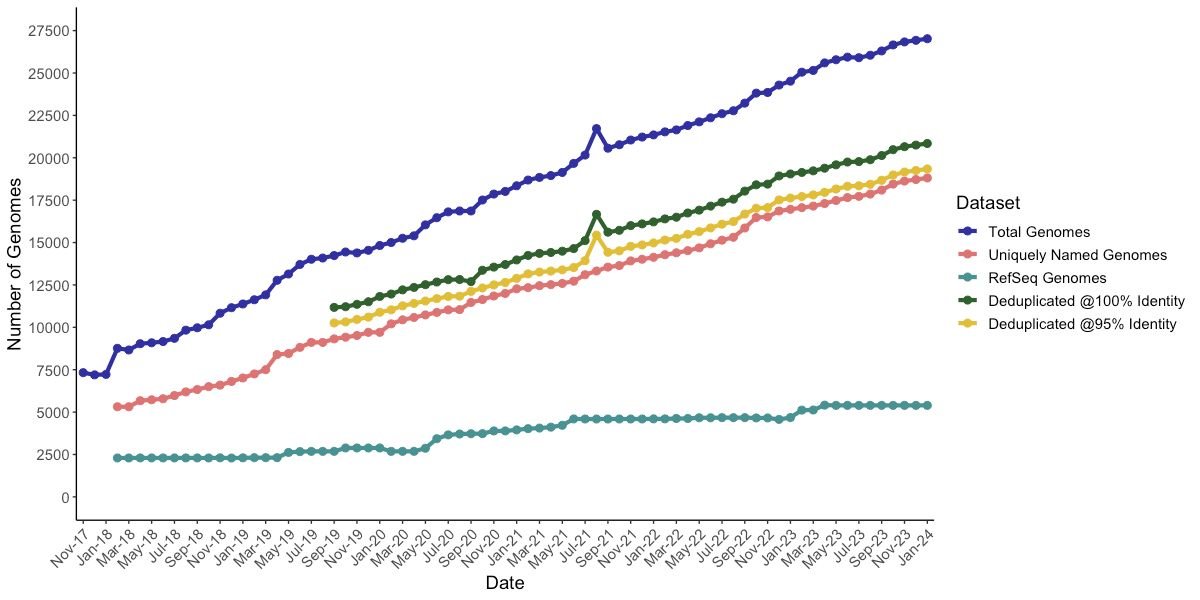
New pre-print with @DovStekelLab, Martha Clokie, Andrew millard and others! We've written a script for the automatic retrieval of bacteriophage genomes from Genbank, and provide useful tools for comparative genomic analysis (1/8) biorxiv.org/content/10.110…







through CLIMB-BIG-DATA I will be running a phage annotation and genomic comparison workshop later in the year (probably april). Poll below, to help us to decide on logistics if you are interested



So happy that Amy Godfrey will be giving a talk at #Microbio22 🎉… She will be giving a talk on bacterial nanosyringes today at 12:15, be sure to check it!!


Learn about GP3 project, tackling Pasteurella infection of salmon with phages, led by Thomas Sicheritz-Pontén .With Bent Petersen, myself, Martha Clokie , and multiple others, including Slawomir Michniewski who just joined the team pasteurella.dk bpetersen.dk/blog youtube.com/watch?v=uODSYS…


Latest from my lab looking at a so called antrax “cross over”strain (B. cereus G9241). Great work by a superb team, Warwick Med School Tom Brooker, Grace Taylor Joyce, Alexia Hapeshi and Shathviga Manoharan. All started off at Bath Uni by Sara. biorxiv.org/content/10.110…


Genome Sequence of SN1, a Bacteriophage That Infects Sphaerotilus natans and Pseudomonas aeruginosa Interesting! 2 bacteria that belong to 2 different classes. Gammaproteobacteria and Betaproteobacteria. I wonder what receptor they share. Sylvain Moineau journals.asm.org/doi/10.1128/mr…

5 undergrads with zero phage experience. 10 weeks later 30+ isolates , 27 phage genomes + TEM images for all, 7 phage with one steps experiments... not to shabby. under expert guidance of Steven Hooton Slawomir Michniewski