
Nucleotide Second Messengers in Bacteria
@spp1879
Mihaela Pruteanu, SPP1879 Managing Coordinator
ID: 984453965239783424
https://www.spp1879.de/ 12-04-2018 15:31:30
1,1K Tweet
498 Followers
512 Following


Check out our newest publication from work funded by DFG public | @[email protected] Nucleotide Second Messengers in Bacteria . Spearheaded by twitterless #AnnaScherhag with excellent supervision by Susanne Zehner

Should I stay or should I go 🦠 lovely new study on bimodal regulation of Pseudomonas aeruginosa biofilm heterogeneity by @Jenal_Lab Biozentrum, University of Basel University of Basel Readcube: rdcu.be/dd8Dg nature.com/articles/s4156…

Do you wonder what nucleotide-based second messengers do in archaea? We do! Read about what is known here: doi.org/10.1093/femsml… Nucleotide Second Messengers in Bacteria Faculty of Biology - University of Freiburg

Now published in an updated form Nature Communications: rdcu.be/de23n Thanks Michael Fuss, Jan-Philip Wiefering, Robin Corey @robincorey.bsky.social, Yvonne Hellmich, Igor Tascon & Joana Sousa at Janet Vonck, Phill Stansfeld & Inga Hänelt labs for your fantastic work! Goethe-Universität Max Planck Institute of Biophysics University of Warwick Nucleotide Second Messengers in Bacteria
Diving into biofilms 🤿 reveals beautiful cellular organization that affects substrate distribution and antibiotic tolerance. Exciting work by Hannah Dayton , our collaborators MinLab@ColumbiaUniv. Anu Janakiraman Jasmine Nirody Raju Tomer & their labs! #microanatomy biorxiv.org/content/10.110…


Finally the last lab day of my PhD has come... Saying goodbye to another beautiful E. coli macrocolony #biofilm 😍 Now on my way to EMBL Heidelberg, looking forward to lots of science at #EESmicrobiology!


Excellent team effort spearheaded by twitterless Simon Brückner together with BangeBalcony and Dominik Begerow, and funded by Nucleotide Second Messengers in Bacteria and GRK2341 "Microbial Substrate Conversion" (MiCon) pubmed.ncbi.nlm.nih.gov/37426605/

Origin and functional diversification of PAS domain, a ubiquitous intracellular sensor | Science Advances science.org/doi/10.1126/sc… Tour de force by Jiawei Xing assisted by Vadim Gumerov and performed at MicrobioOhioState If you have a PAS domain, please, do send it to us 😎!

The VAAM - Vereinigung f. Allg. u. Ang. Mikrobiologie conference has started: Regine Hengge delivers the Schlegel lecture! Nucleotide Second Messengers in Bacteria


Happy to share our latest work on #cdiAMP signaling & natural #competence via #ComFB protein, a novel #cdiAMP receptor (widespread in bacteria 🦠) Great collaboration w/ Sonja-Verena Albers & Shamphavi Sivabalasarma Supported SPP2389 Bacteria! Beyond the single cell! Nucleotide Second Messengers in Bacteria @sfb1381 Faculty of Biology - University of Freiburg Controlling Microbes to Fight Infections biorxiv.org/content/10.110…

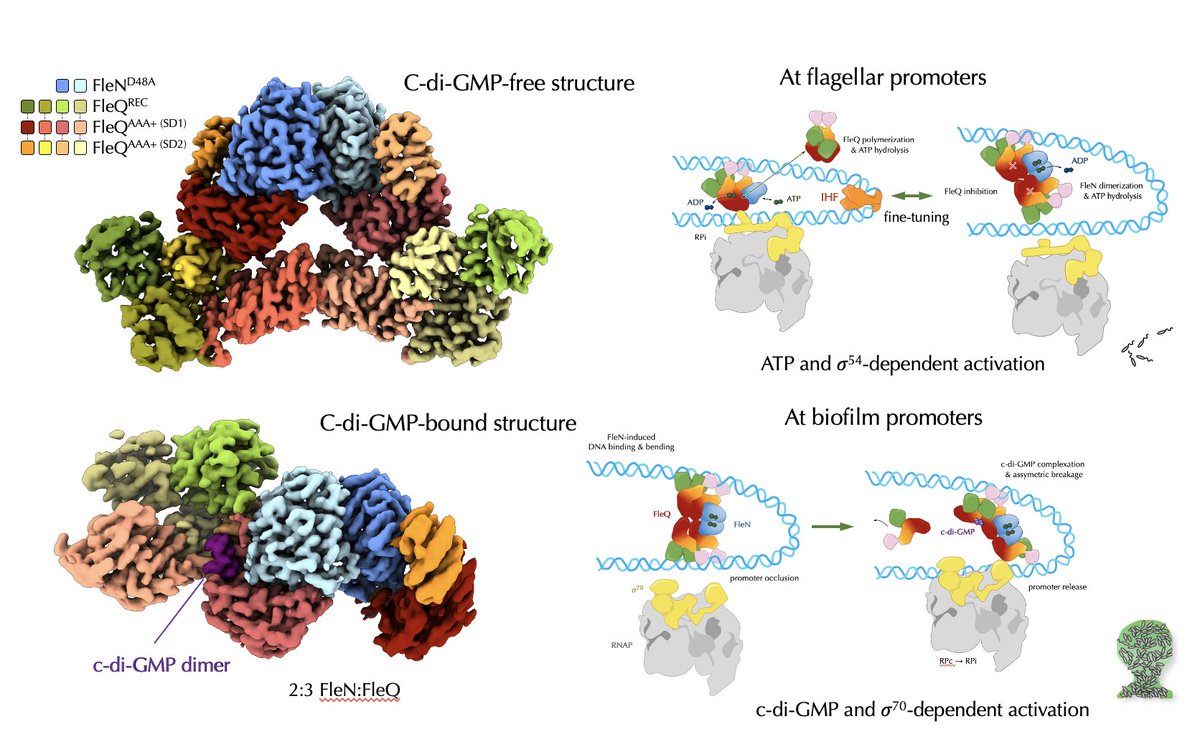
Apparently our latest paper just came out 🤩! Structures of the P. aeruginosa FleQ-FleN master regulators reveal large-scale conformational switching in motility and biofilm control. PNASNews European Research Council (ERC) Institut Européen de Chimie et Biologie University of Bordeaux CNRS Biologie. pnas.org/doi/abs/10.107…


Wow: C-di-AMP accumulation disrupts glutathione metabolism and inhibits virulence program expression in Listeria monocytogenes: biorxiv.org/content/10.110… #cdAMP Nucleotide Second Messengers in Bacteria

A must read for #cdiAMP community & for Nucleotide Second Messengers in Bacteria followers ;-)

Posttranscriptional modification of #cdiAMP synthase DisA tunes down it's activity a good read for Nucleotide Second Messengers in Bacteria community journals.asm.org/doi/10.1128/mb…

👉 From dusty shelves toward the spotlight: growing evidence for Ap4A as an alarmone in maintaining RNA stability and proteasis! pubmed.ncbi.nlm.nih.gov/39216180/ Nucleotide Second Messengers in Bacteria #Ap4A #subtiwiki


Great study about the link between c-di-AMP metabolism and cell wall integrity: Nucleotide Second Messengers in Bacteria
