Sangram Kadam
@sangram__kadam
Postdoctoral Associate @schlicklab @nyuniversity | PhD @LabRanjith @iitbombay
Website: sangramkadam.github.io
ID: 1015176525010702337
06-07-2018 10:11:59
31 Tweet
96 Followers
223 Following

Interesting work from our lab alumnus Srivastav Ranganathan on RNA-binding protein condensates and emergence of fibrillar structures using coarse-grained simulations.


Our work, modeling the high fidelity inheritance of histone modifications post replication, using ideas from information theory, is published in PLOS computational biology. journals.plos.org/ploscompbiol/a… Ranjith Padinhateeri

This reminds me of a video that we - @sangramjkadam Navin Sridhar and I made a looong ago in our undergraduate years using a stop motion animation! Just a camera setup in the hostel room. Here is the link.. youtu.be/ukuiGlHx-wI


Congratulations to Prof. Ranjith Padinhateeri for being elected as a Fellow of the Indian Academy of Sciences, Bengaluru.


Our work is out in Nature Communications. Read the paper if you want a quantitative description of chromatin polymer or plan to simulate chromatin. We answer some fundamental questions. Details below. @sangramjkadam Kiran K. Vinoth Shuvadip Dutta mithunmittir (1/10)

Happy to share our latest preprint where we explore how the clustering of enzymes can improve metabolic fluxes even when the enzymes themselves are far from "perfect". biorxiv.org/content/10.110… Work done with Junlang Liu and Prof. Eugene Shakhnovich.




"How high-impact papers from Indian researchers are shaping science". nature.com/articles/d4158… One of these from MajiLab (Protein Engineering and Neurobiology Lab) IIT Bombay. Samir Samir K Maji has pioneered work on aggregation & phase separation of proteins associated with neurodegenerative diseases. Biosciences & Bioengineering, IIT Bombay

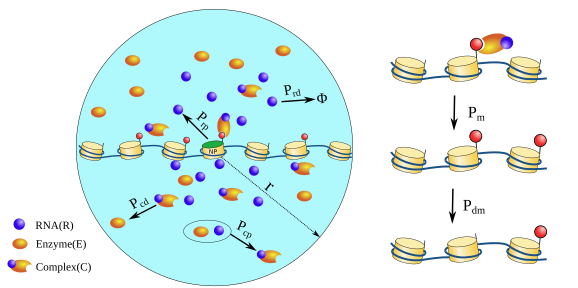
Our latest paper to wrap up the year. We propose that intersegmental jumps that leverage looped conformations of chromatin can lead to efficient search. Read the preprint at: biorxiv.org/content/10.110… Led by Shuvadip Dutta & Adarshkrishnan. Fantastic collab. with Ranjith Padinhateeri (1/4)

Samir K Maji Biosciences & Bioengineering, IIT Bombay IIT Bombay Congratulations Samir... great going.


Excited to be a part of #BPS2024 and present my work. for details, have a look at our preprint: biorxiv.org/content/10.110… mithunmittir Ranjith Padinhateeri #Philadelphia Biophysical Society