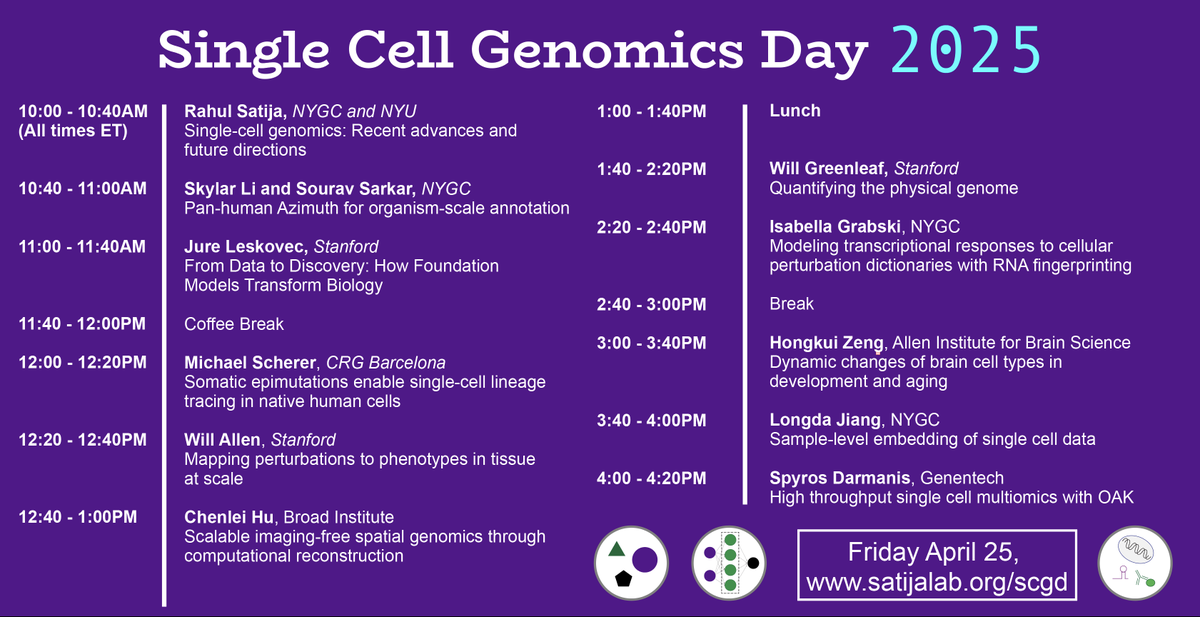
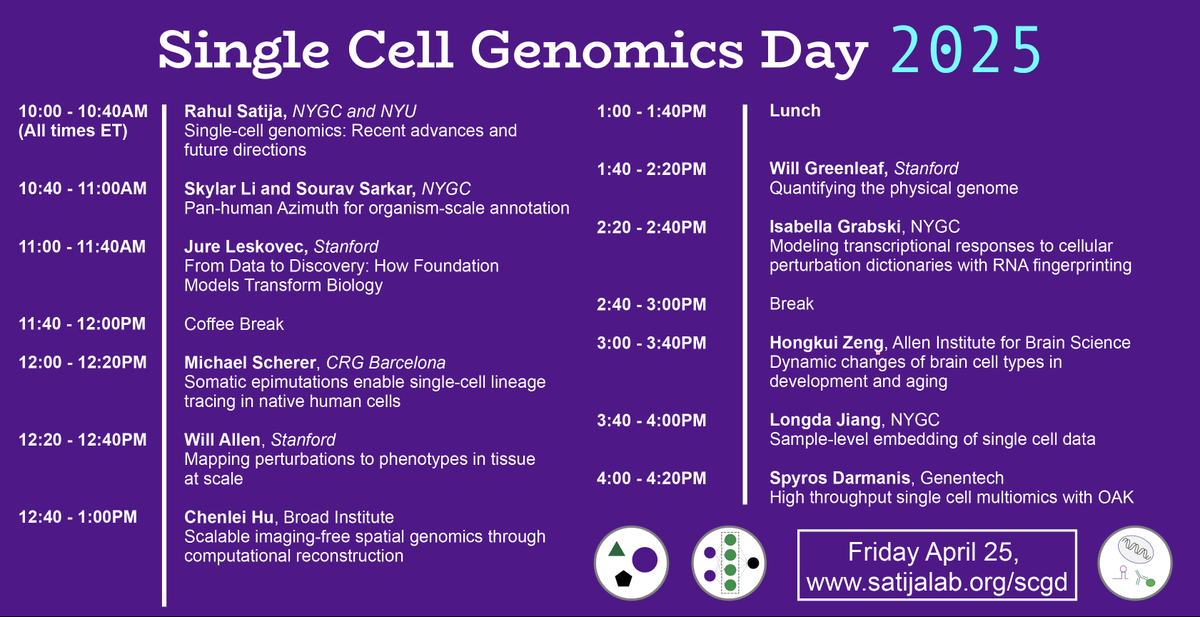
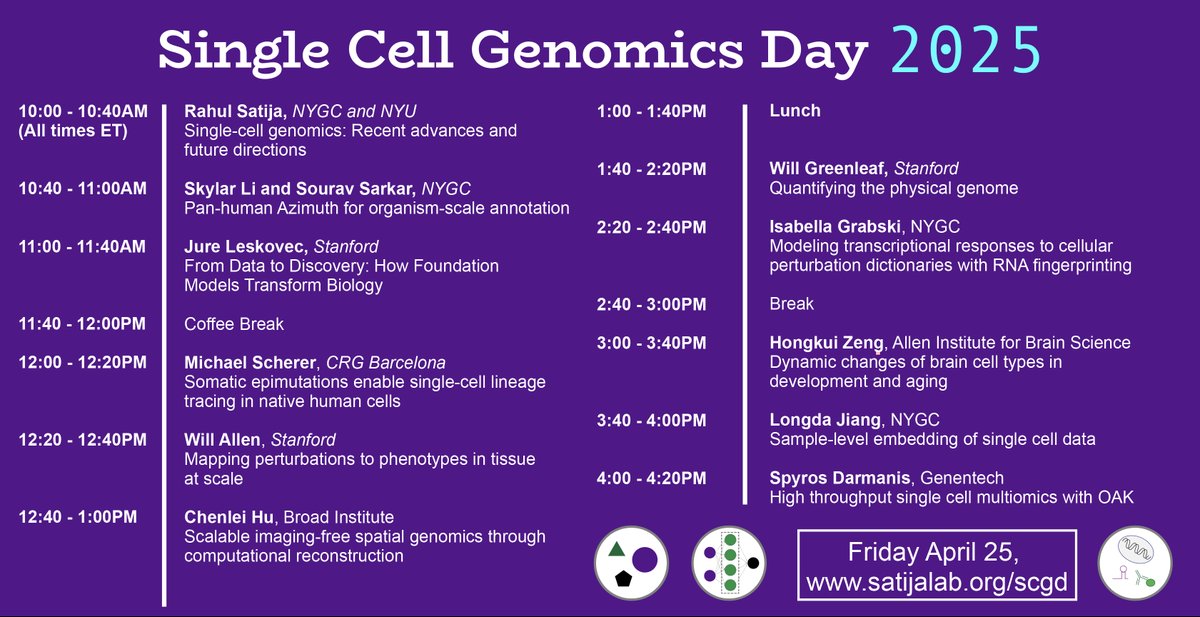
Rahul Satija
@satijalab
Core Member, New York Genome Center; Professor, Biology, NYU
ID: 76515242
http://www.satijalab.org 23-09-2009 01:13:40
900 Tweet
27,27K Followers
355 Following



Cool work from spyros darmanis that performs combinatorial-based superloading of 10x Genomics for scRNA-seq and multiome assays. Interesting idea to add the 10x barcode first, then the second barcode later: nature.com/articles/s4146…


In collaboration with Reuben Saunders, Jonathan Weissman's Lab, and Xiaowei Zhuang, we are very excited to release Perturb-Multi: a platform for pooled multimodal genetic screens in intact mammalian tissue. Check it out! biorxiv.org/content/10.110…

Finally out in Science Magazine! Utilizing the cost-effective EasySci to profile 21 million single-cell transcriptomes across five life stages and diverse sexes/genotypes, we reveal aging as distinct, development-like transitions, with dramatic cell population changes in specific


We're incredibly excited to share our new method, Paired-Damage-seq, out Nature Methods today! This technique allows us to jointly analyze oxidative DNA damage and SSBs with RNA, all at the single-cell level (1/5). nature.com/articles/s4159…


Excited to share our exploration Nature Methods into new horizons opening up in the era of low cost sequencing! 🔥 Deeper sequencing at WGS scale for PPM sensitivity of tumor informed MRD 🔥Duplex sequencing at WGS scale for plasma only exploration of ctDNA Check out thread 👇




Excited to share our first anti-aging intervention study, led by brilliant Zehao Zehao Zhang, Alex Alex Epstein, and Chloe Chloe Schaefer from our lab at Rockefeller University! With a cost-effective spatiotemporal analysis pipeline, we identified the aging-associated cell