Sergey Melnikov
@sergeyvmelniko1
Group Leader @UniofNewcastle pushing our way into the unknown. An avid lover of hibernation and (ironically) coffee.
ID: 1158784482620710912
http://melnikovlab.com 06-08-2019 16:58:44
232 Tweet
356 Followers
259 Following

Our latest study on the structural basis of deciphering AUA codon by isoleucine tRNAs was published at NatureStructMolBiol back-to-back with the paper by Gagnon Lab Matthieu Gagnon. Congratulation Akiyama-san! doi.org/10.1038/s41594…

Out just now The EMBO Journal: our cryo-EM structure of the native RanGTP-induced γ-TuRC-capped microtubule, revealing a surprisingly asymmetric minus end architecture with direct implications for microtubule regulation and modulation during spindle assembly. embopress.org/doi/full/10.10…

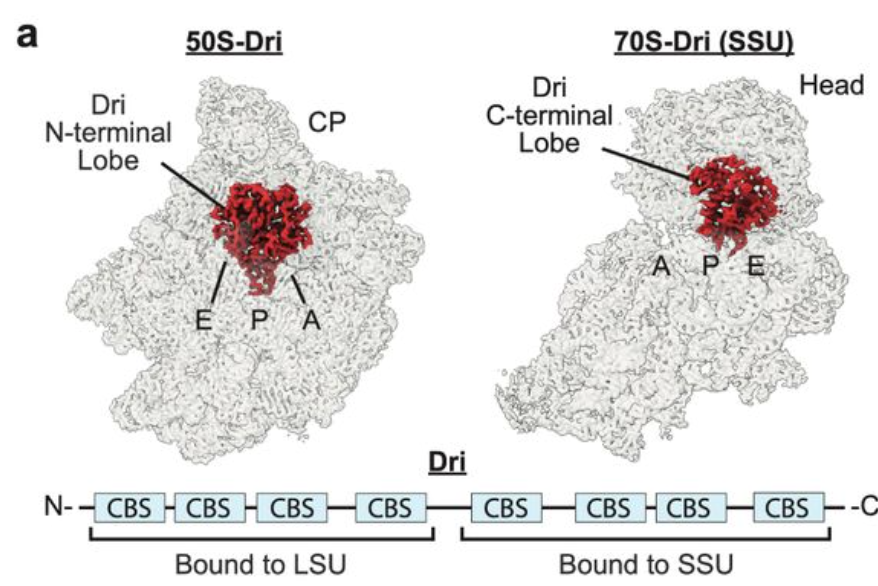
Our cryoEM of CBS filament conformations out in Nature Communications. Began decade ago when TJ McCorvie (@allostericstate on ☁️) purified oligomers. Fab collab Andre LB Ambrosio Sandra Dias seeing them in cells. Huge thanks Centre for Medicines Discovery Pfizer Inc. HCU Network America Wellcome for support x.com/NatureComms/st…

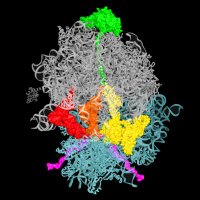
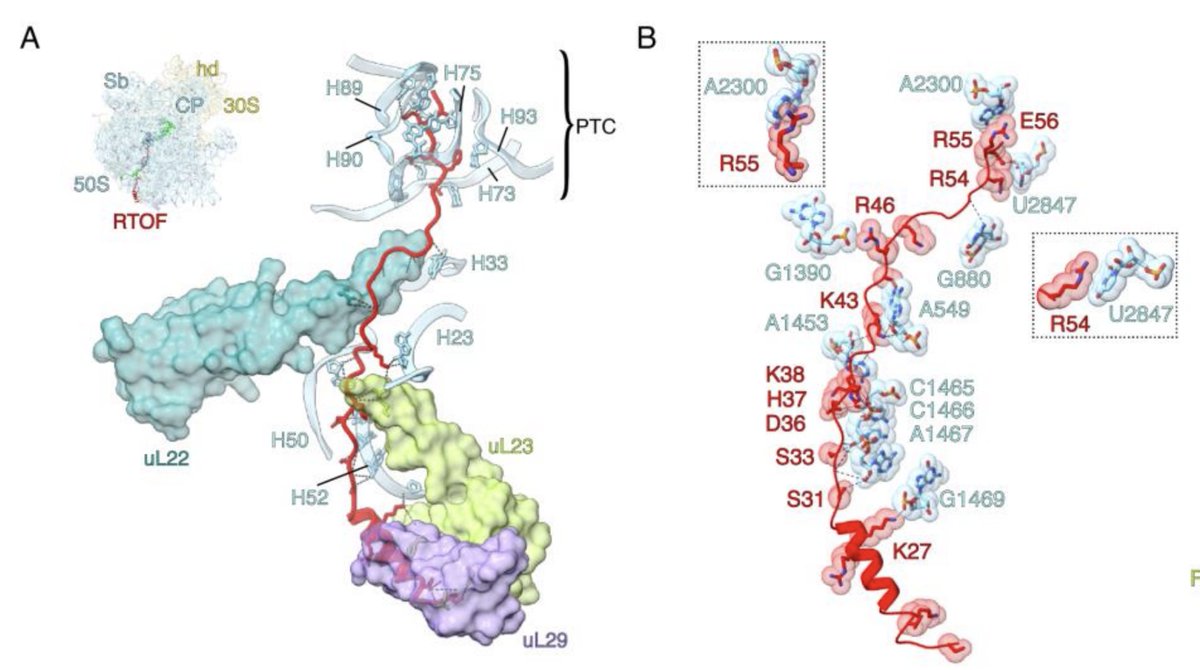
Angiogenin controls angiogenesis (et plus) by nicking tRNAs. But it’s barely active alone. We discovered how 80S ribosome unlocks Ang’s active site and “brings in” tRNAs. Congrats to Anna Loveland and collab. w/Allan Jacobson! nature.com/articles/s4158…
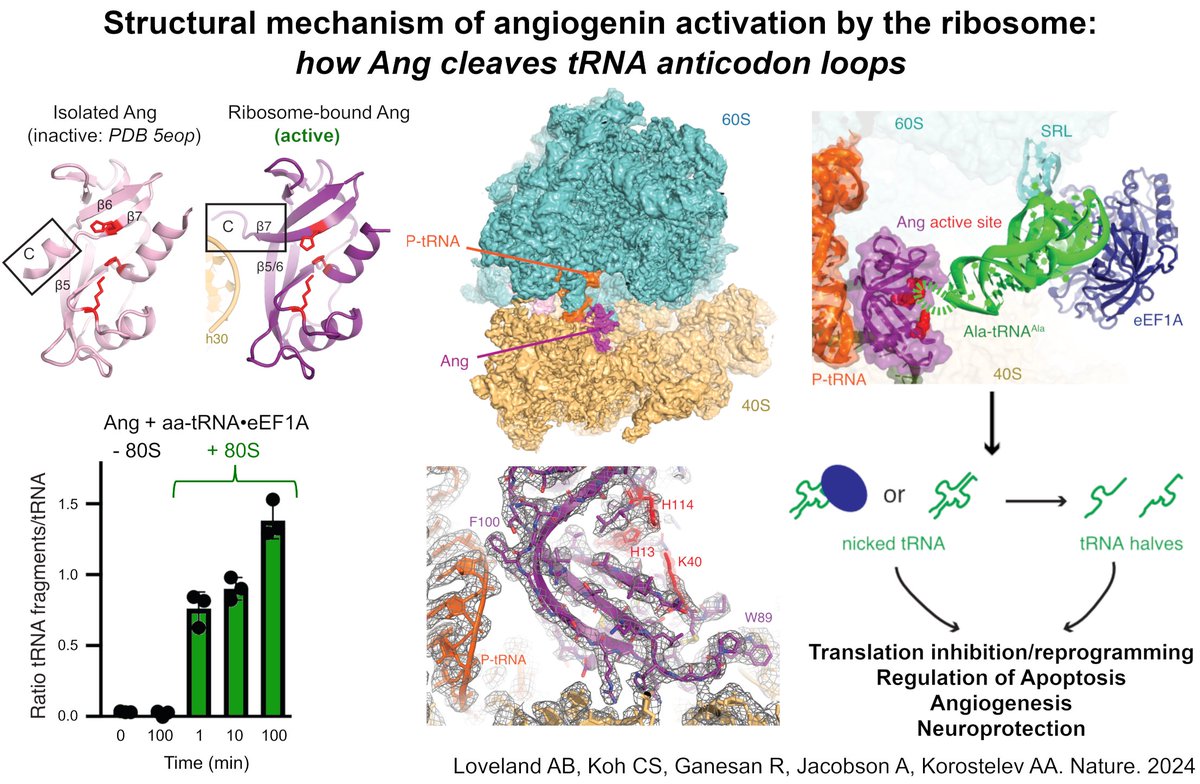









Studying ribosomes? Read this work by Sergej Djuranovic and slavica pavlovic djuranovic—as it presents one of the most rapid and cost-efficient methods to isolate ribosomes from rare organisms or small biomass quantities 💪biorxiv.org/content/10.110…
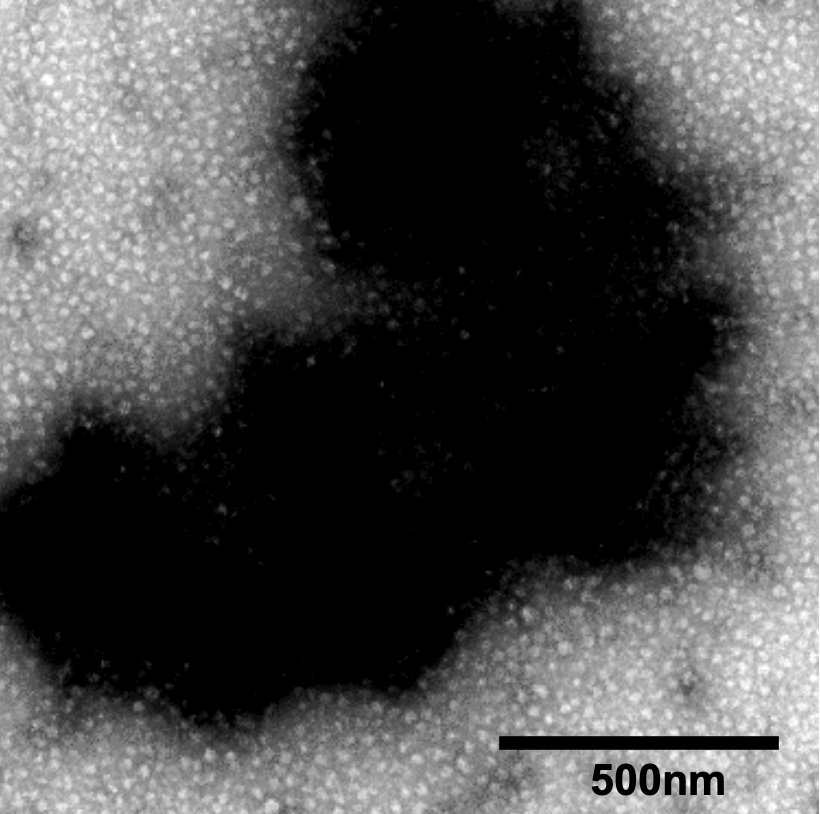

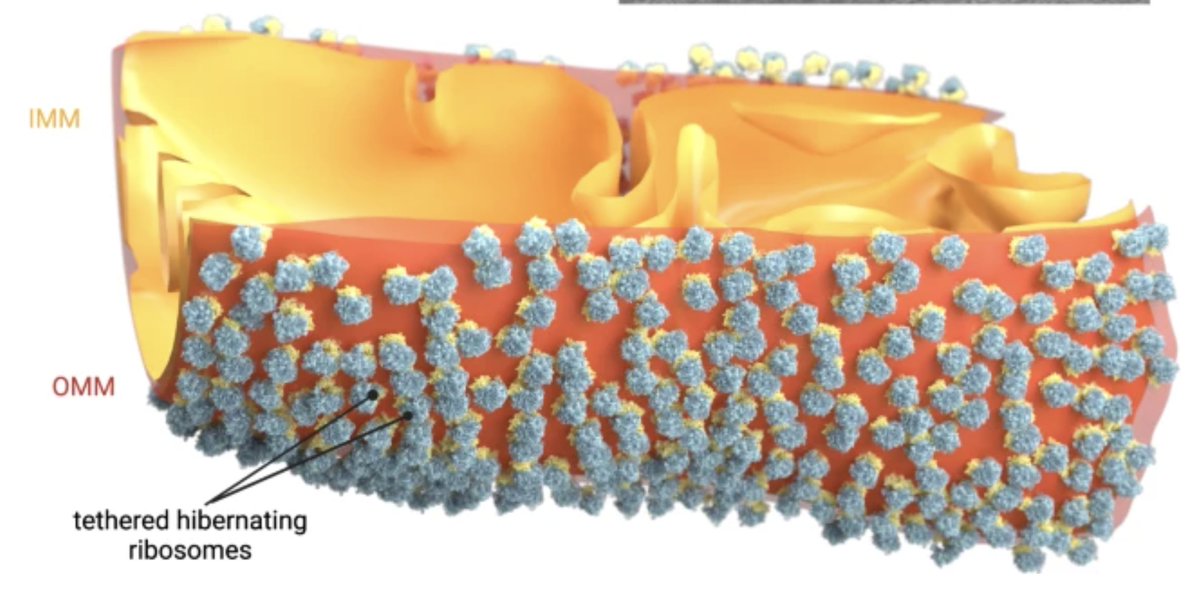
Hibernating ribosomes have a complex social life in starved yeast cells: they assemble into intricate clusters and bind to mitochondria, illustrating how ribosomes hibernate in their natural habitat. More—in this fascinating study by Ahmad Jomaa: nature.com/articles/s4146…