Sid Annaldasula
@sidannaldasula
Interested in genomics! Researcher at BIMSB and Botanischer Garten. Bioinformatics Masters Student at FU Berlin. Fulbright Germany '19. 🐧 🧬
ID: 1310521913081462785
28-09-2020 10:09:20
32 Tweet
53 Followers
113 Following

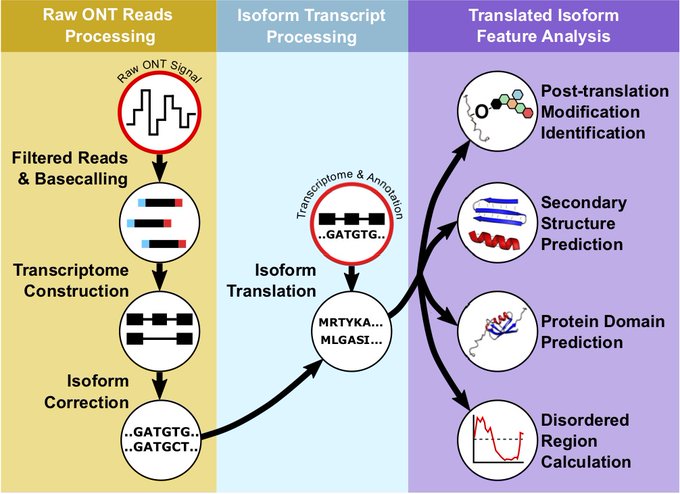
Excited to share IsoTV, a tool I developed at the Mayer Laboratory along with Martyna Gajos! IsoTV predicts and visualizes functional features of transcript isoforms. Read more about it here: isotv.readthedocs.io



The preprint from the Singapore Nanopore Expression Project (SG-NEx) is online! The SG-NEx data is a systematic resource for Oxford Nanopore long read RNA-Sequencing, including direct RNA, cDNA, and PCR cDNA with matched short read data and spike-in RNAs. 1/n biorxiv.org/content/10.110…


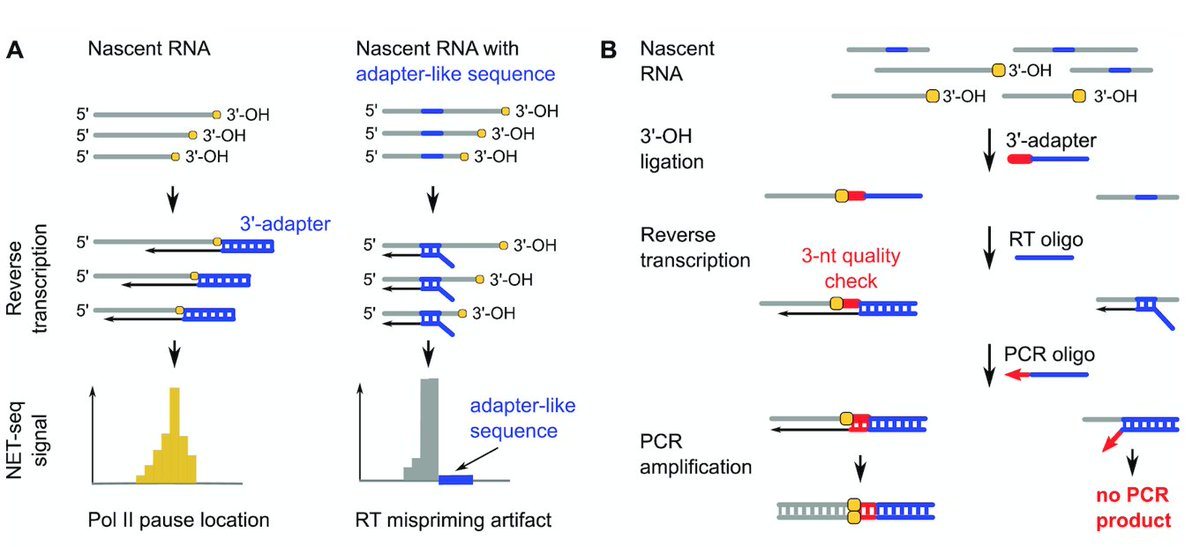
I'm very happy to finally share Annkatrin Bressin's & my work on an unexpected role of BRD4 in transcription termination! authors.elsevier.com/c/1dUFc3vVUPGK… Congrats & thanks to all involved, especially Andreas Mayer, Olga Jasnovidova, PhD, David Meierhofer!

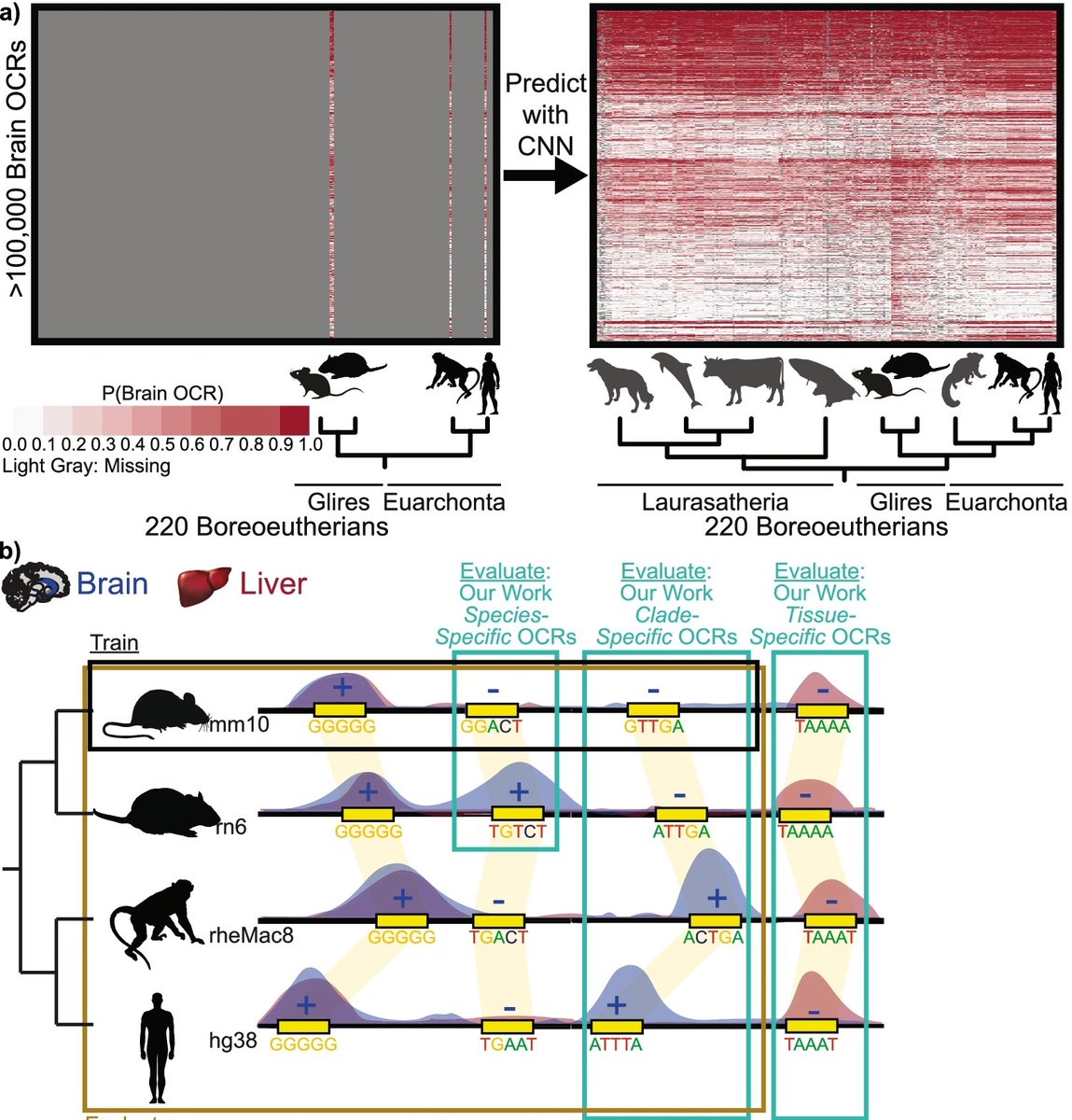
Ever wanted to intersect cell types, epigenomics, ML, human genetics, and addiction together? Check out new publication: doi.org/10.1523/JNEURO… Thanks to much work from Chai and coauthors Alyssa Lawler Andreas Pfenning morgan wirthlin Easwaran Ramamurthy Michael Kleyman 🇺🇦


So excited to announce the first preprint from the Haghverdi lab is out!👑 Check out the wonderful work of Valérie Marot-L. , Brigitte Bouman and Fearghal Donaghy “Towards reliable quantification of cell state velocities” doi.org/10.1101/2022.0…

Check out our latest review on the R language! Federico Giorgi, @Notorious_D_ and I had a lot of fun writing it and I hope you will enjoy reading it! 😊 mdpi.com/1606768

Our manuscript, summarizing the second part of my PhD work, is out! We present Association Plots, a tool for visualization of #markergenes, adapted to #SingleCell data. sciencedirect.com/science/articl… Big thanks to Clemens Kohl, Bita Sokhandan Fadakar, and Martin Vingron VingronLab!



📍Posting live from the grounds of Charité - Universitätsmedizin Berlin in Mitte! We @compcancer are showcasing the importance of computational tools in cancer research at the Long Night of Science @lndwberlin. Sid Annaldasula Javier Marchena-Hurtado


Compound-SNE is out in Bioinformatics: a tSNE-based embedding method for visual alignments of single-cell data in embedding space that is applicable across different samples, data modalities and experimental timepoints. Congrats, Colin Cess & Laleh Haghverdi! academic.oup.com/bioinformatics…