Thomas TOPILKO
@ttopilko
Scientist at Lundbeck 🇩🇰. Neuroscience, Spatial biology & Python 🐍. Previously @GubraBiotech 🇩🇰 | @RockefellerUniv 🇺🇸 | @InstitutCerveau🇫🇷.
ID: 1148936478636093440
10-07-2019 12:46:17
274 Tweet
184 Followers
224 Following







How cool is this? Michal Lipinski from Broad Institute presenting stacked #Illumina spatial slides of the mouse brain with subcellular transcriptome resolution #AGBTGM

Hiroki R. Ueda すいしぇ/Susaki EA Our 3D whole-brain single-neuron atlas is now available on GitHub! This atlas integrates spatial information of every cell in the mouse brain within the Allen Brain Atlas CCFv3. Our paper links it with spatial transcriptomics dataset. github.com/OrganismalSyst…


Advances in neuroscience have yielded massive, rich datasets from various analysis techniques and modalities. In a Nature Neuroscience perspective, researchers underscore the importance of an integrative approach to render a more complete picture of the brain. nature.com/articles/s4159…
🥼 Janelia researchers led by Boaz Mohar (@[email protected]), Nelson Spruston, Luke Lavis & karel svoboda Allen Institute have developed a new imaging tool that maps brain-wide changes in neuronal connections. 🧠 🔗 janelia.org/news/new-imagi…

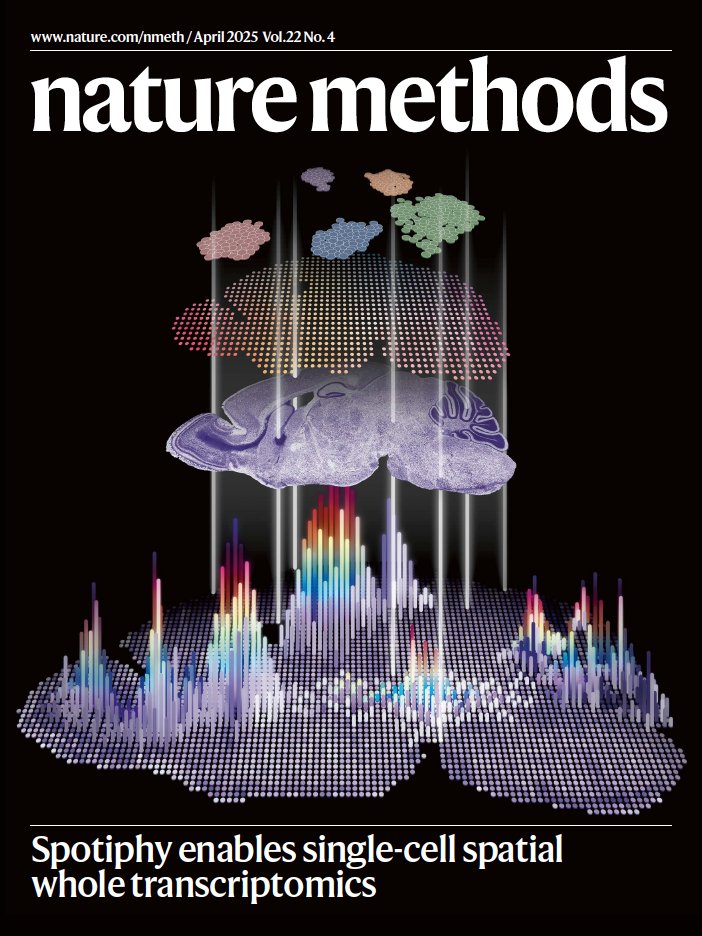
Our April issue is live! 🌧️🌷 nature.com/nmeth/volumes/… On the cover, Spotiphy projects scRNA-seq and histological image data onto the spatial transcriptome. Artwork by Jiyuan Yang St. Jude Research. Paper: nature.com/articles/s4159…

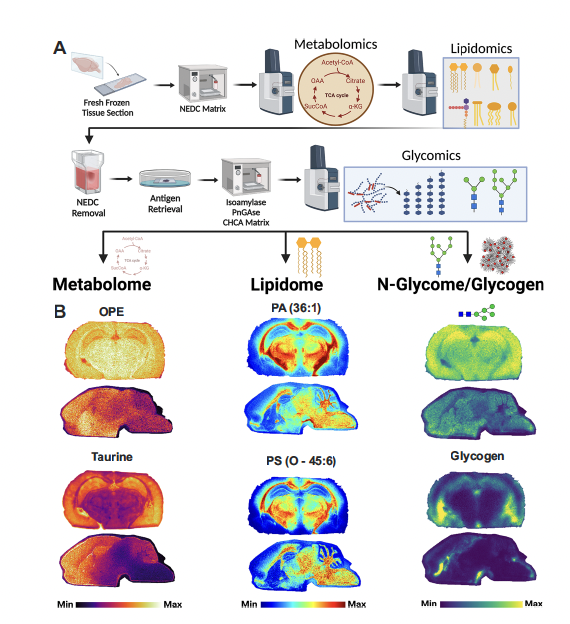
🚨 New in Nature Communications: We present a spatial multi-omics platform that maps the metabolome, lipidome, and glycome from a single brain section. Combining MALDI imaging with our computational tool Sami, we reveal metabolic diversity in health and Alzheimer's disease. 🔗


The MOSAIC reconfigurable microscope by Eric Betzig et al is finally out! MOSAIC integrates multiple advanced imaging techniques including light-sheet, label-free, super-resolution, and multi-photon, all equipped with adaptive optics: biorxiv.org/content/10.110…