Theo Hirsch
@tz_hirsch
Cancer Genomics Researcher @Inserm, zucmanlab.com @CRCordeliers | Liver cancer evolution in children and adults | Bulk, single-cell, spatial | Mosaicism
ID: 1934656308
https://cvscience.aviesan.fr/cv/11954/theo-hirsch 04-10-2013 16:05:34
312 Tweet
191 Followers
362 Following


PAPER OUT!! 🥳🎉💃🏻🕺🏻💃🏻🕺🏻 A redefined InDel taxonomy provides insights into mutational signatures Nature Genetics Brainchild of the amazing Gene C. Koh Tweetorial below!! 👇 1/n nature.com/articles/s4158…

Stromal mutations occur, but hard to prove they are functional. In our new paper with Trey Ideker’s lab and led by Dr. Mohita Tagore and Maayan Baron, we show that we can find these by looking for mutations across a “class” of genes rather than single genes: nature.com/articles/s4158…

Thrilled to share our latest study just out in nature 😄A major milestone in our collaboration with Anirban Maitra Led by brilliant Guangsheng Pei Jimin Min MD Anderson Cancer Center nature.com/articles/s4158…

🚨Thrilled to share our latest work: “Tumor-Infiltrating Clonal Hematopoiesis” NEJM - we investigated how age-related blood mutations impact cancer when they infiltrate tumors, with Elsa Bernard Maria Zagorulya, Elli Papaemmanuil, PhD Charles Swanton + more nejm.org/doi/full/10.10…






Fantastic paper from Ben Creelan, MD lab showing subclonal neoantigen loss in tumours progressing on TIL therapy highlighting importance of targeting clonal neoantigens - watch this space for Darwin2 trial reported ESMO - Eur. Oncology by Crispin Hiley Nicholas McGranahan

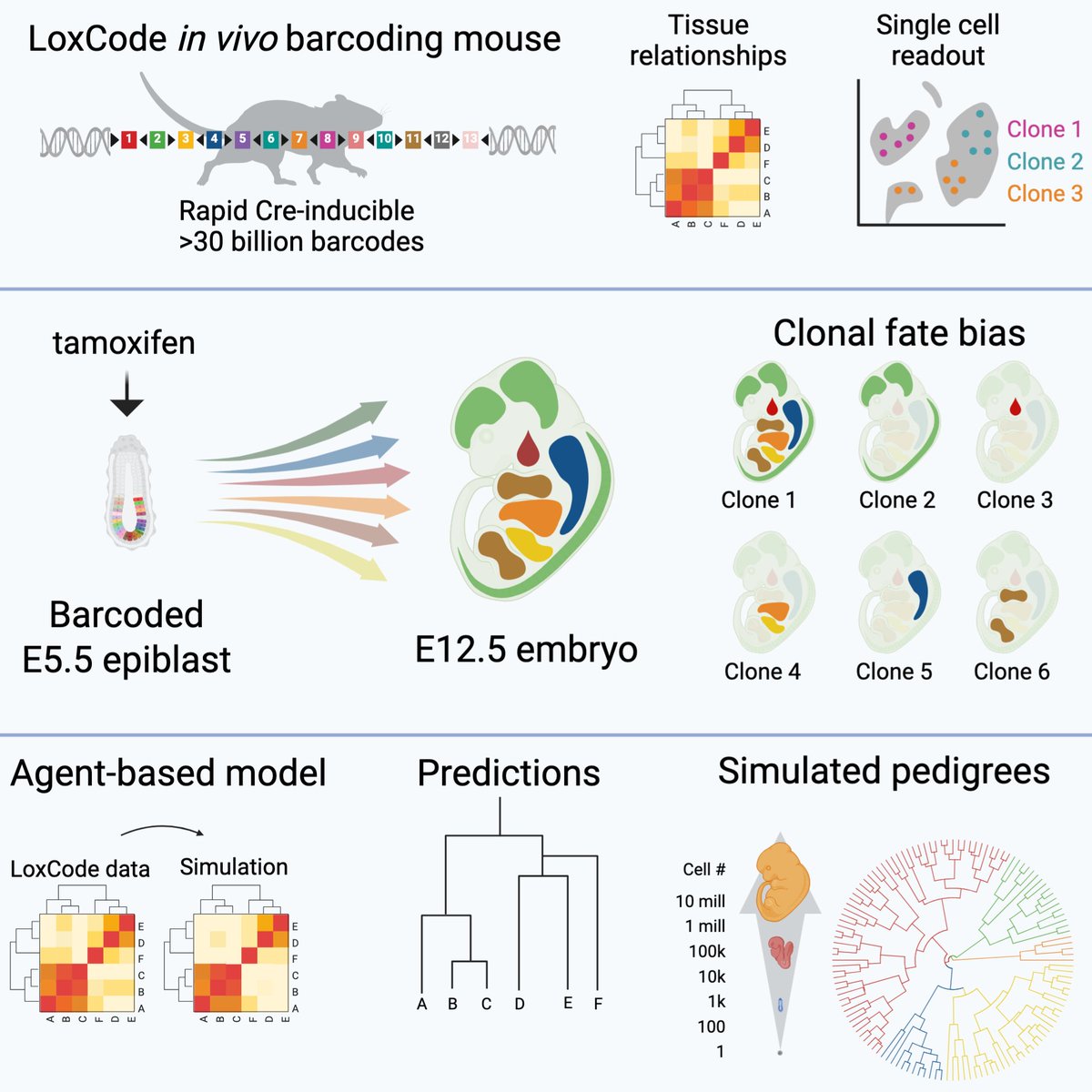
A triumph of perseverance from twitterless Tom Weber, Christine Biben and the team, our in vivo barcoding "LoxCode mouse" used to resolve epiblast fate to fetal organs is finally published in Cell and available to import through The Jackson Laboratory sciencedirect.com/science/articl…

I’m excited, relieved, and honored to announce that my paper describing non-canonical mitotic mechanisms in the early mouse embryo is out in Science Magazine ! (link at end of 🧵)

In Nature Genetics we report that base editing of trinucleotide repeats (TNRs) reduces somatic repeat expansions in Huntington’s disease (HD) and Friedreich’s ataxia (FRDA)—in patient-derived cells and in vivo—a collaboration with the Mouro Pinto lab. drive.google.com/file/d/1on7kdy… 1/14



Thank you so much to the Fibrolamellar Cancer for the invitation to the fibrolamellar scientific summit in Greenwich! It was an honor to meet this great community of scientists, physicians and patients. A lot of fruitful discussions and I hope some new collaborations!