
Thirumalai Lab @ UT Austin
@thirumalaiarmy
Thirumalai group account.
ID: 1413640564025724937
https://sites.cns.utexas.edu/thirumalai/home 09-07-2021 23:26:05
51 Tweet
274 Followers
25 Following


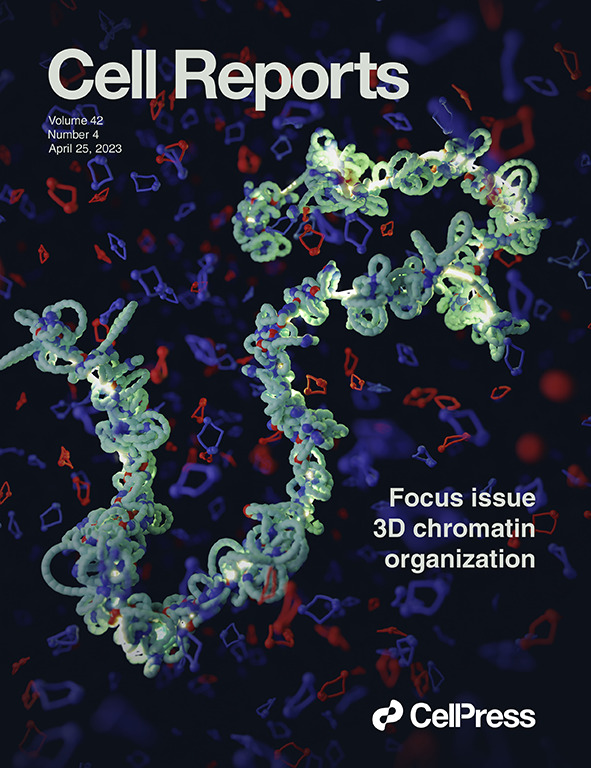
Our research was featured on the cover of the current issue of Cell Reports. The picture is an artistic interpretation of how mitotic #chromosomes are built by #condensin molecules. Made in Blender 🔶 using the #molecularnodes package developed by Brady Johnston.


We are happy to share our recent work on PNASNews (pnas.org/doi/10.1073/pn…). Thanks to Dave Thirumalai, Hung Nguyen, Naoto Hori and Thirumalai Lab @ UT Austin . We established a link between the folding landscapes of the monomers of CAG repeats RNA and their propensity to self-associate.

We introduce a new sequence-specific force-field (christened as COFFEE) for simulations of DNA-protein complexes. COFFEE reproduces many experimental results (SAXS, NMR), and accurately describes the nuances of nucleosome dynamics.Thirumalai Lab @ UT Austin biorxiv.org/content/10.110…

Wondering how glycans affect the ACE2-RBD binding in SARS-CoV-2 infection? Our new Thirumalai Lab @ UT Austin paper in Biophysical Journal highlights that the interactions of glycans in ACE2 receptors are largely entropic rather than enthalpic! Check out more here ➡️ authors.elsevier.com/a/1h7lh1SPT42Q8

Congratulations to Debayan Chakraborty for his position at IMSC chennai as an assistant professor. Dr. Chakraborty will be joining later this month. We wish him a succesful independent career, and stay tuned for exciting biophysics coming from his lab.


Congratulations Dr. Davin Jeong for successfully defending her PhD!! Davin Jeong studied interphase chromosome structures with MD simulations and data-driven methods. Read up on her work here biorxiv.org/content/10.110…


We had a farewell party for Hung Nguyen who starts his independent career at UB Dept of Chemistry. Best wishes for Hung and his future!







In eLife - the journal: Structural basis for the preservation of a subset of topologically associating domains in interphase chromosomes upon cohesin depletion doi.org/10.7554/eLife.…. #3Dgenome. Another eLife paper from Thirumalai Lab @ UT Austin Davin Jeong @anyuzx.


Excited to share our new paper: "Emergence of cellular nematic order is a conserved feature of gastrulation in animal embryos" with Thirumalai Lab @ UT Austin, @Jbwallingford, Mark Peifer, Margot Kossmann Williams, Jessica, Robert J. Huebner. Read it here: biorxiv.org/content/10.110…. #devbiol #nematic



My study on glycan-glycan interactions is now published in J. Am. Chem. Soc.! We found that uncharged N-glycans show long-range pair repulsion, which varies logarithmically (⬅️unexpected analogy w/ star polymers!). We highlighted the role of water in the repulsion. pubs.acs.org/doi/10.1021/ja…




![Sucheol Shin (@sucheol_shin) on Twitter photo RT plz! APS Focus Session "Genome Organization & Subnuclear Phenomena" is coming back at the 2025 Global Physics Summit! We invite the abstracts of latest developments in the 4D genome using experimental, theoretical, and/or computational methods, from all-level scientists! [1/4] RT plz! APS Focus Session "Genome Organization & Subnuclear Phenomena" is coming back at the 2025 Global Physics Summit! We invite the abstracts of latest developments in the 4D genome using experimental, theoretical, and/or computational methods, from all-level scientists! [1/4]](https://pbs.twimg.com/media/GZnz4xaWgAU7dO9.jpg)