Tim Lohoff
@tim_lohoff
Life Science Venture Capitalist @ForbionLifeSci | Ph.D. in Stem Cell Biology & Medicine from @Cambridge_Uni @ReikLab
ID: 827086228805779456
02-02-2017 09:28:18
94 Tweet
273 Followers
445 Following

Congratulations Diljeet Gill @aledjohnparry and all other authors 🥳. One of the most exciting projects from the lab showing that transient reprogramming leads to rejuvenation of human fibroblast! Check out the preprint 👇:
You wanna build muscles? 💪🏼 At least in vitro we know how 😉 Check out our latest paper eLife - the journal investigating forces during muscle development! Huge shout out to Gilbert Lab and @JoergEnderlein for making it so beautiful! elifesciences.org/articles/60145


E8.5 seqFISH data from Tim Lohoff/Shila Ghazanfar/Cai Lab is on MouseGastrulationData devel now! Count matrices, annotation, spot locations, cell segmentation masks, imputed Tx-wide counts - the whole shebang. Give it a go for methods work or dev bio! bioconductor.org/packages/devel…


Really excited to share our paper on Nature Genetics about the impact of mitochondrial metabolism on nuclear DNA methylation. Thanks to European Research Council (ERC) ,Armenise Harvard Foundation, @Telethonitalia, Università di Padova for the amazing support. go.nature.com/39CU7Ur

Did you ever wonder if there are any biomarker for brain injuries 🧠? Read the nice commentary from the talented Ruslan Rust to learn more!👇🔬🙂

Job alert! We're looking for three postdoctoral researchers to lead exciting projects investigating the #epigenetic regulation underlying lineage specification during #development. Apply here: babrahaminstitute.livevacancies.co.uk/#/job/details/… #academicjobs IHEC_Epigenomes



Impressive and important work 🐭 Congratulations to Ruslan Rust and all other authors! 🤩🥳 check it out 👏💥

Our work "Integration of spatial and single-cell transcriptomic data elucidates mouse organogenesis" is now out at Nature Biotechnology ! Congratulations to all the authors! nature.com/articles/s4158… #seqfish #spatial #singlecell #transcriptomics



Incredibly happy to share that our work, co-led with Tim Lohoff is now available online at Nature Biotechnology 🎉 Feeling very grateful to everyone who was involved in this project 🙏 Explore the data here: marionilab.cruk.cam.ac.uk/SpatialMouseAt…

Our work is out in Science Magazine! Using integrated spatial genomics technology, we have extensively investigated the single-cell nuclear architecture across cell types in the mouse brain cortex with a great team Cai Lab Guo-Cheng Yuan Shiwei Zheng 郑诗蔚 #seqFISH science.org/doi/10.1126/sc…

Wow 🤯 Congratulations Felix Lansing and team 🎉 Amazing work. Looking forward to following the path of designer-recombinases towards the clinic 🩸💪🏼 Keep the exciting work up! 🍀 Check out the great tweetorial below 👇

Great work Ruslan Rust and team! 🙌🐭

Happy to see another piece of my PhD work published in Genome Biology! 🥳 Amazing collaboration with @RArgelaguet and Stephen Clark 💪#DreamTeam Big thanks to everybody involved Reik Lab Cambridge Stem Cell Institute Cambridge University Babraham Institute! Finally, check out the great tweetorial below 👇:

New preprint! 🥳 biorxiv.org/content/10.110… Led by @AledJohnParry & @ChristelKrueger we asked why #DNAmethylation and oxidation often target overlapping genomic regions at the exit of #pluripotency. Great collaboration with Tim Lohoff, Steven Wingett & @stefanschoenfe1. 🧵👇

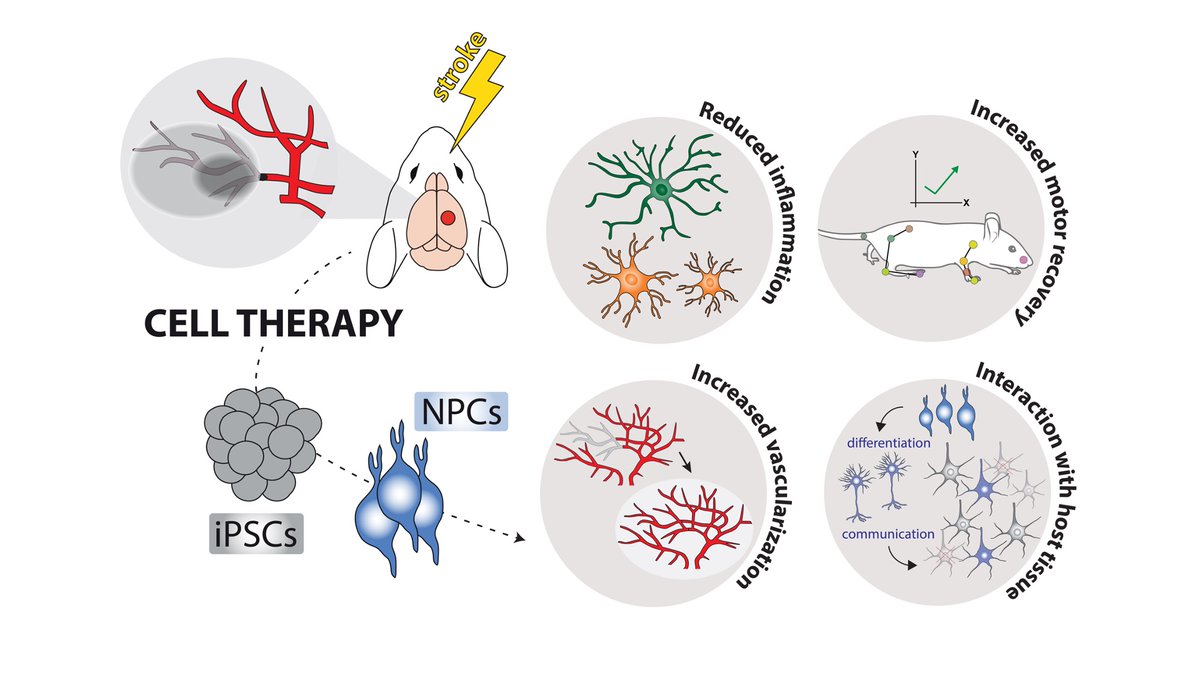
New preprint! 🚨Keck Medicine of USC Institute for Regenerative Medicine We started with the simple question: What happens when we transplant stem cells into a mouse model of #stroke, and what are the interactions between graft and host? Took us only 5 yrs to find out😀: biorxiv.org/content/10.110… 1/10 🧵