
Tong Ding
@tongding99
PhD student in Computer Science at @hseas
ID: 1684227655225475072
26-07-2023 15:42:40
17 Tweet
76 Followers
108 Following


Excited to present UNI - a general-purpose self-supervised visual model for #CPath pretrained using 100M+ images across 100K+ WSIs! Co-led with Tong Ding Max Lu @DFKW_MD Faisal Mahmood Harvard Medical School Summary: bit.ly/3EzEFr0 Preprint: arxiv.org/abs/2308.15474 1/


⚡️🔬📣Excited to share our two new Nature Medicine articles, we develop computational pathology foundation models, 1. UNI, a self-supervised computational pathology model trained on 100 million pathology images from 100k+ slides. 2. CONCH, a vision-language model for
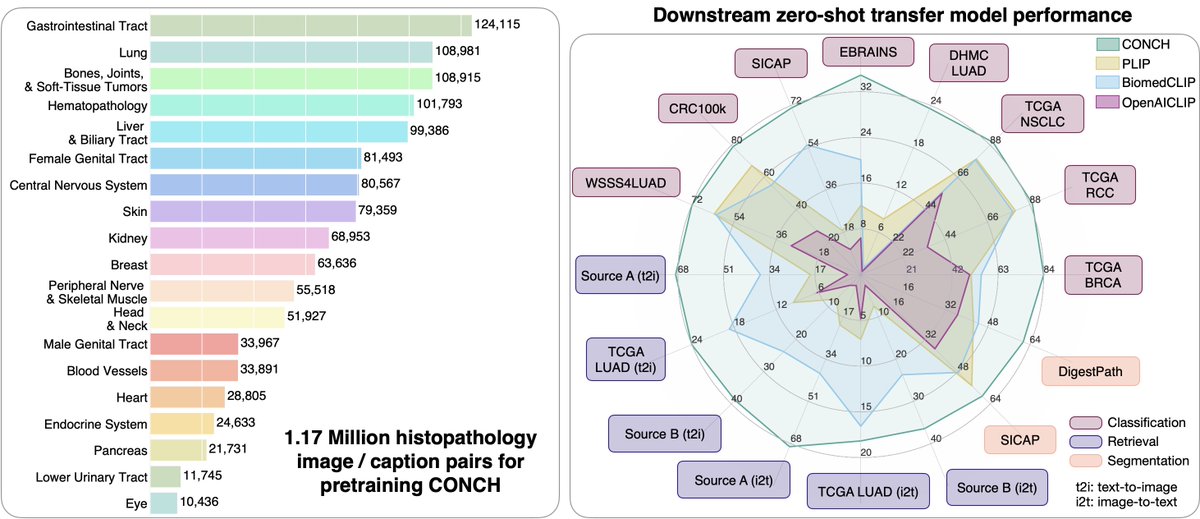

Two remarkable new papers Nature Medicine on foundation models #AI for pathology nature.com/articles/s4159… 100,000 whole slide images w/ >100 million path images nature.com/articles/s4159… Multimodal of ~1.2 million images and text pairs Faisal Mahmood Richard J. Chen Max Lu @DFKW_MD


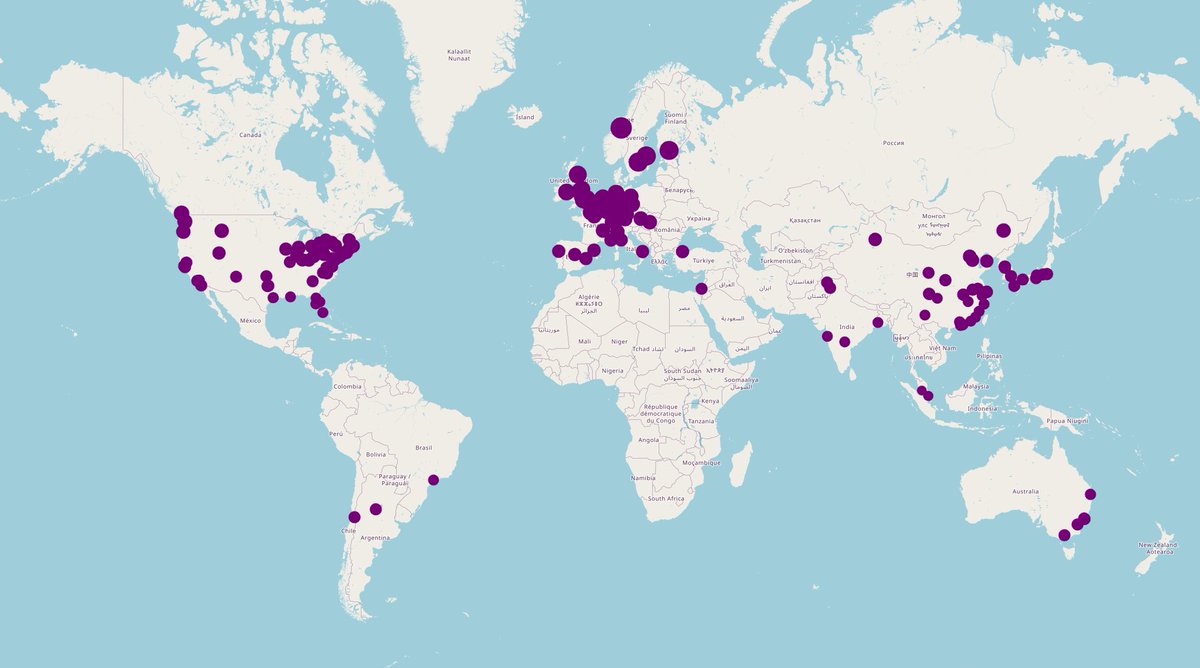
Its been a month since we released UNI and CONCH, a vision only and a vision-language foundation model for computational pathology w/ Nature Medicine articles. The models have been downloaded over 233k times on @HuggingFace we are analyzing the early impact, see where people are



Excited to announce (*4 months late😅) that I am #phdone! Crossing this finish line would not be possible without the support and collaboration of many colleagues and mentors, and also without Faisal Mahmood ! With Faisal Mahmood , Guillaume Jaume , Andrew H. Song,



Excellent work from Gabrielle Campanella, Thomas J. Fuchs Icahn School of Medicine at Mount Sinai! Benchmarking 8 pathology foundation models including UNI, Virchow and Prov-GigaPath on large scale datasets for clinical diagnosis and biomarker discovery and providing insights into data scaling laws and














