WESTPA Software
@westpasoftware
Moved @westpasoftware.bsky.social. Highly scalable software for weighted ensemble simulations with any dynamics engine, including MD and systems biology.
ID: 1090273536927772672
https://westpa.github.io/ 29-01-2019 15:40:42
56 Tweet
264 Followers
83 Following



Beyond coarse-grained modeling: Seconds-timescale protein conformational change measured using Cu(II)-based EPR and weighted ensemble MD simulations. Lillian Chong Congratulations to Xiaowei and @AnthonyBogetti on this work, now published in Protein Science: onlinelibrary.wiley.com/doi/abs/10.100…


Using WE simulations and experiments, The Chong Lab and the Horne lab explore the atomistic folding pathways of protein mimetics pubs.rsc.org/en/content/art…

Register now for the free WESTPA Workshop Pitt Chemistry, June 20-21. Explore rare event simulations like protein folding, ligand binding, & chemical reactions with our open-source software. Perfect for those using AMBER, NAMD, GROMACS, etc. Reg info: forms.gle/6gJF6jJ72iexbD…

Another great BPS with both vintage and fresh WESTPA developers/users from the Lillian Chong Rommie Amaro Daniel Zuckerman Terra Sztain, PhD and Ahn Labs!


It's finally out! Outstanding work by @atbogetti Matthew Zwier using WESTPA Software AMBER md





New work on protein design... 🚨S2 immunogen design using advanced simulation WESTPA Software WE MD--> opening mechanism--> design--> stabilized protein--> cryoEM structure + immunology Xandra Nuqui Lorenzo Casalino Jason McLellan Kartik Chandran nature.com/articles/s4146…


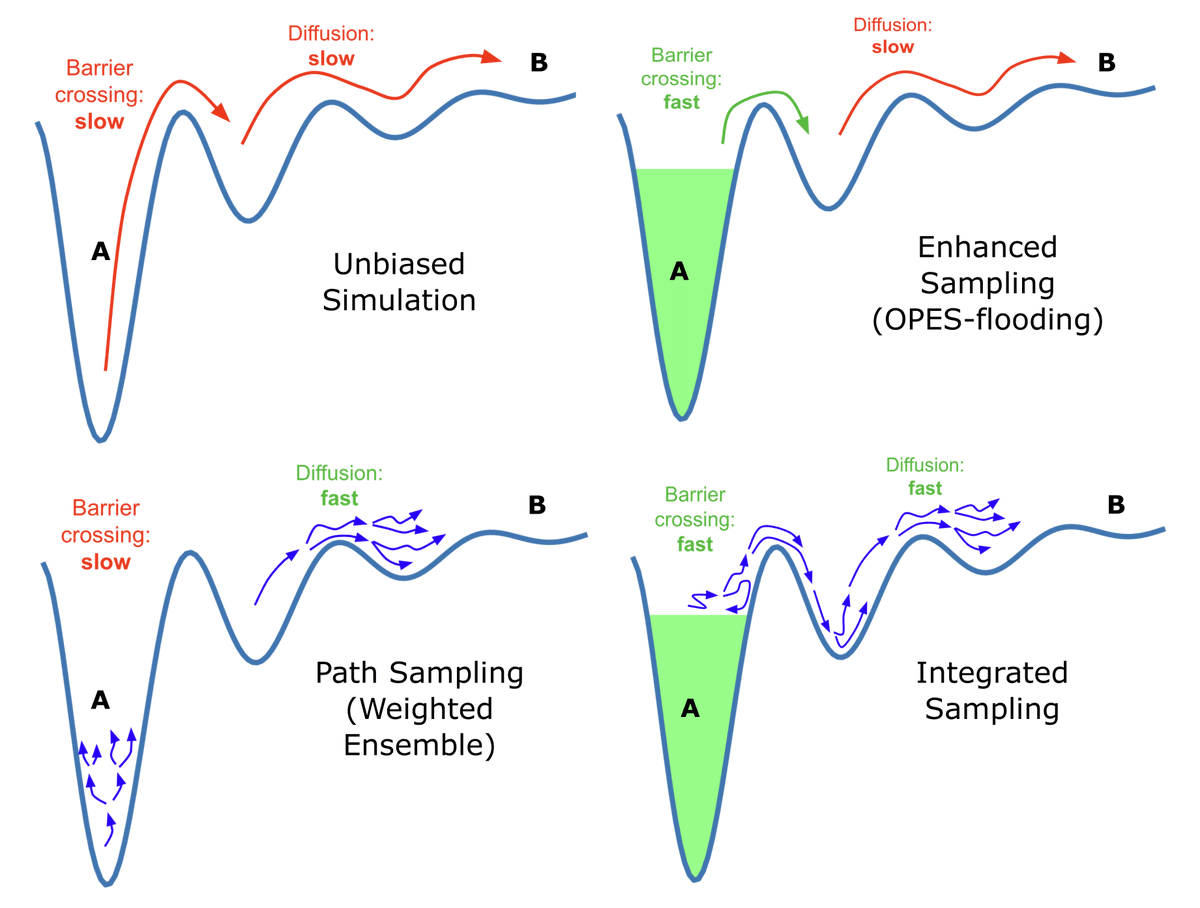
Just out in The Journal of Chemical Physics! Integrating Weighted Ensemble (WESTPA Software) with OPES-Flooding for accurate and efficient calculation of rare-event kinetics involving barrier crossing and diffusion (e.g. protein folding, ligand binding) UO Chem and Biochem #compchem doi.org/10.1063/5.0239…