Yang-Hong Dai
@yanghongdai1
Radiation Oncologist|Oxford University|Department of Oncology|Graph neural network
ID: 835721775640039425
26-02-2017 05:22:53
320 Tweet
54 Followers
198 Following














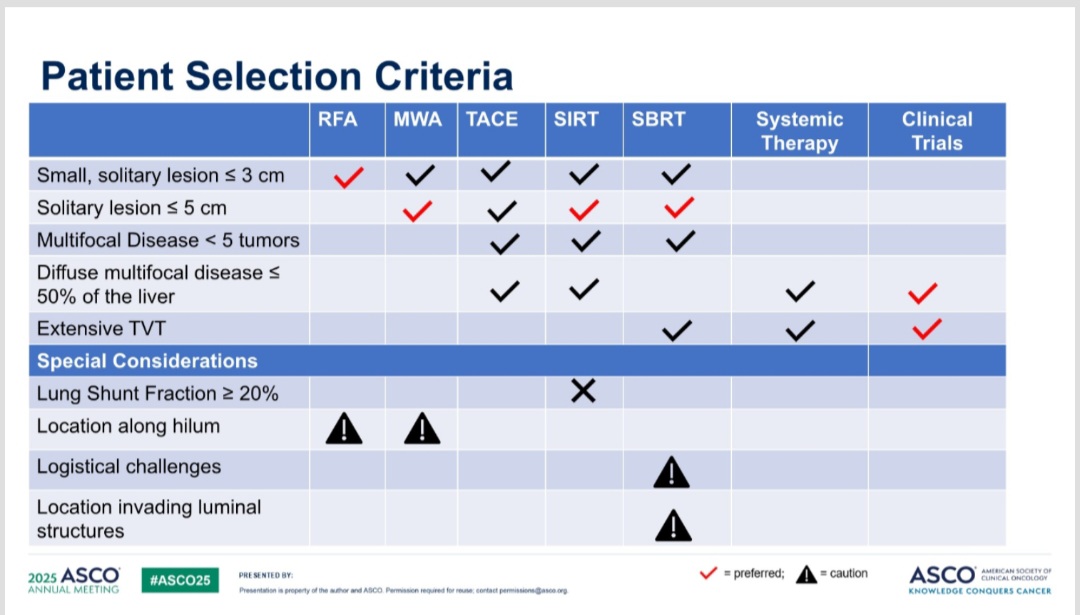
How to select pts with HCC for various local therapies? RFA ,MWA , TACE , SIRT , SBRT . Useful for clinical decision-making. Taken from #asco25 ASCO Do u agree ? Yakup Ergün Dr. Nina Niu Sanford Jeff Ryckman OncoAlert Kate Sears