Zechen Chong
@zechenchong
#bioinformatics Associate Professor at UAB Informatics Institute
ID: 128105896
https://sites.uab.edu/chonglab 31-03-2010 03:30:31
383 Tweet
174 Followers
658 Following

Looking to show off your new variant finder, interpreter, visualizer? Submit to our #bibm2022 workshop on Computational methods to characterize genomic variants using high-throughput sequencing data. Papers due Oct. 10 (Don't worry. I'll remind you) combioconference.github.io/bibm2022-works…



Check out this short course for #primarycare providers to learn more about #raredisease. 7/27 at 3ET It’s led by an amazing group of experts and will be very practical! Registration link in the original post #MedTwitter National Organization for Rare Disorders (NORD)


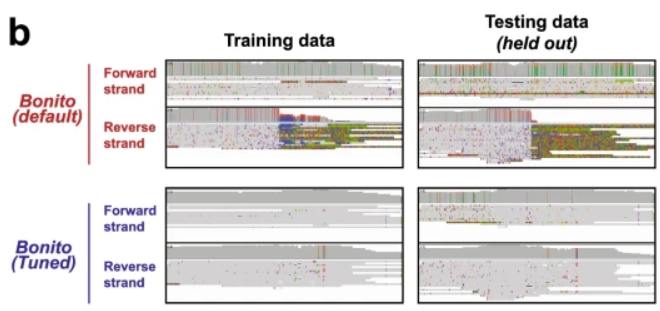
Study of basecalling errors in nanopore sequencing of telomeres, from Kar-Tong Tan, Slevin, Meyerson & Heng Li. Telomeres in many organisms are frequently miscalled, but tuning the basecalling models improves this, with minimal impact in other regions genomebiology.biomedcentral.com/articles/10.11…





Applications open for a PhD in Swanton Lab The Francis Crick Institute to study the impact of environmental pollution on lung cancer initiation/promotion- join an incredible team+ institute - please apply or retweet Science and Innovation at Cancer Research UK UCL Cancer Institute crick.ac.uk/careers-study/…

Fast-Higashi is featured on the cover of Cell Systems. Diverse cell types (marbles) can be characterized by single-cell 3D chromatin structures. Fast-Higashi identifies latent correlation of different cells (connections among marbles). cell.com/cell-systems/f…





Great to see that our work on gene fusion detection in long-read cancer transcriptome appears in the first issue of Cancer Research aacrjournals.org/cancerres/arti…

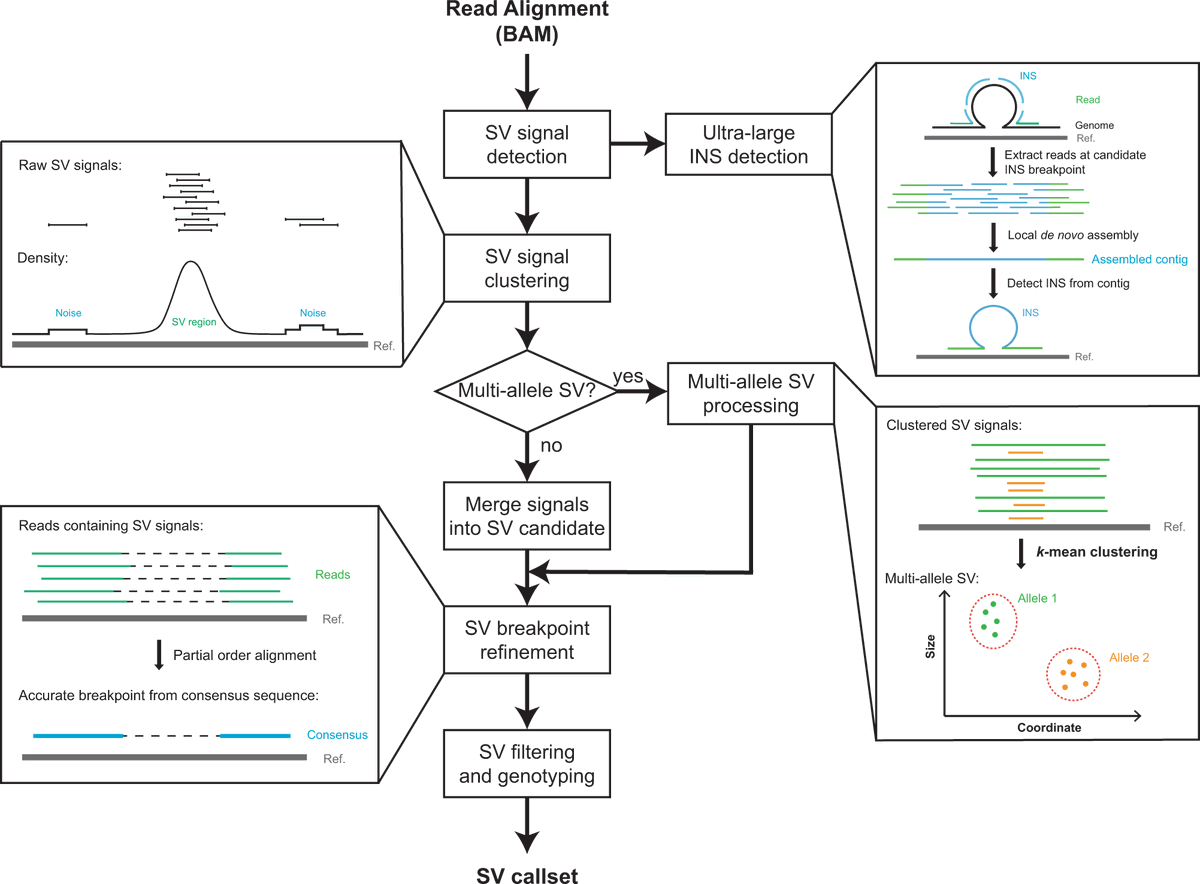
Our work, DeBreak, on long-read SV detection is online now Nature Communications: nature.com/articles/s4146…. Welcome to test it out. #longreadsequencing #SV #structuralvariation

Recently published: Zechen Chong and colleagues develop DeBreak, an algorithm for comprehensive and accurate #StructuralVariant #SV detection in #LongReadSequencing data across different platforms #Genomics #CancerResearch Cancer at Nature Portfolio nature.com/articles/s4146…

A personal update while working on #RNA23 badge... Excited to announce that Su Lab will open its door UAB Genetics in July! We are recruiting at all levels for anyone who is interested in RNA modifications, gene regulation, NGS analysis and cancer biology. (1/4)