
Christian Zimmerli
@zimmerlichris
DoYouEvenTilt?
ID: 1053745834737745920
20-10-2018 20:32:39
31 Tweet
112 Followers
108 Following


Do you know how the architecture of the nuclear pore is linked to its assembly and turnover in living cells? Out in @nature now! Congratulations to @Matteoffm Christian Zimmerli Jan Kosinski Vasileios Rantos Boris Pfander @EMBL Max Planck Institute of Biophysics Max Planck Institute of Biochemistry (MPIB) (1/6) nature.com/articles/s4158…

Didn't know this could have turned in such an emotional event. I feel happy, proud, lucky and so much grateful! B. Turonova, Thorsten Müller, Marina Lusic, Simone Mattei, B. Müller and H-G Kräusslich! Special thanks to Martin Beck, Vojtech Zila, Christian Zimmerli and @Matteoffm

What if building large multicomponent protein complexes becomes as likely as winning the lottery? Our analysis is now online. Thanks everyone! Max Planck Institute of Biophysics MPI for Brain Research EMBL Hummer Lab Kiran Raosaheb Patil Erin schuman
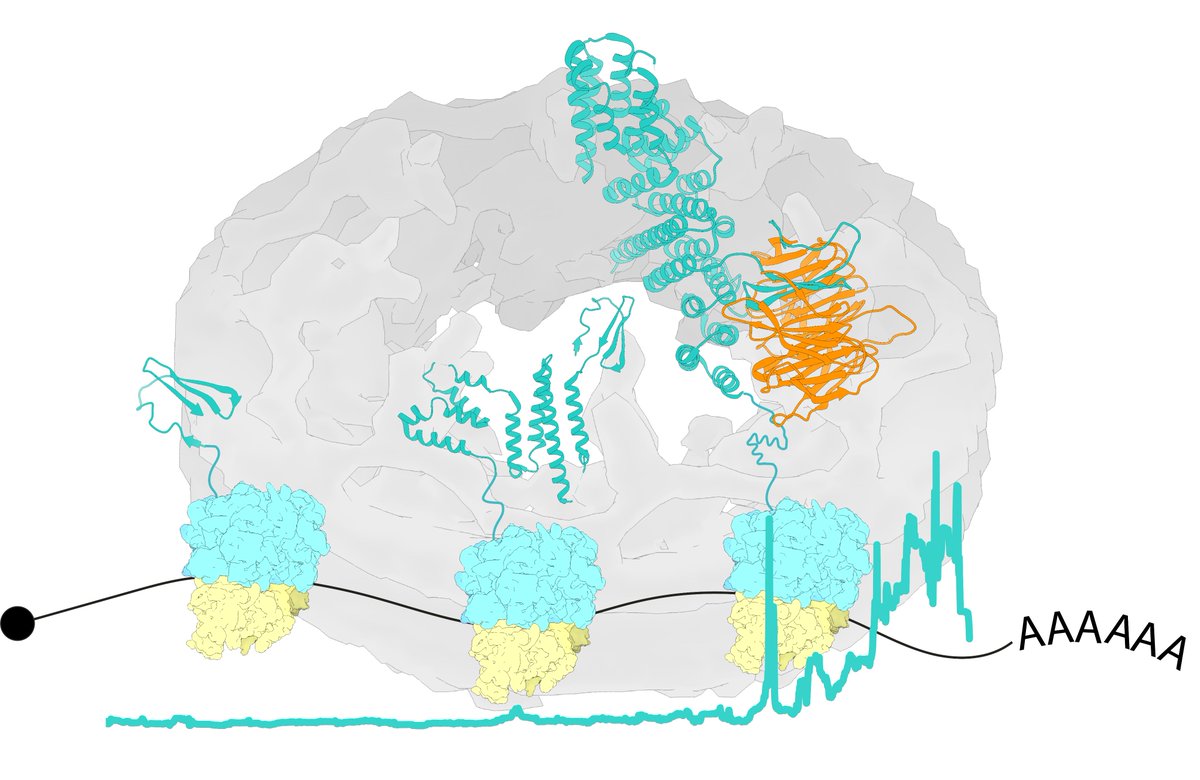

So happy to finally share this with the🌍!! We've come a long way, thanks to everyone involved! Special thanks to @Matteoffm, Jan Kosinski, Beck Laboratory, MahamidLab and Hummer Lab. It was a blast! #TeamTomo #CryoEM

Vasileios Rantos, Agnieszka Obarska, and I did the modeling based on in-cell cryo-electron tomography maps from Christian Zimmerli and @Matteoffm from Beck Laboratory. Modeling as always using our Assembline protocol embl-hamburg.de/Assembline/

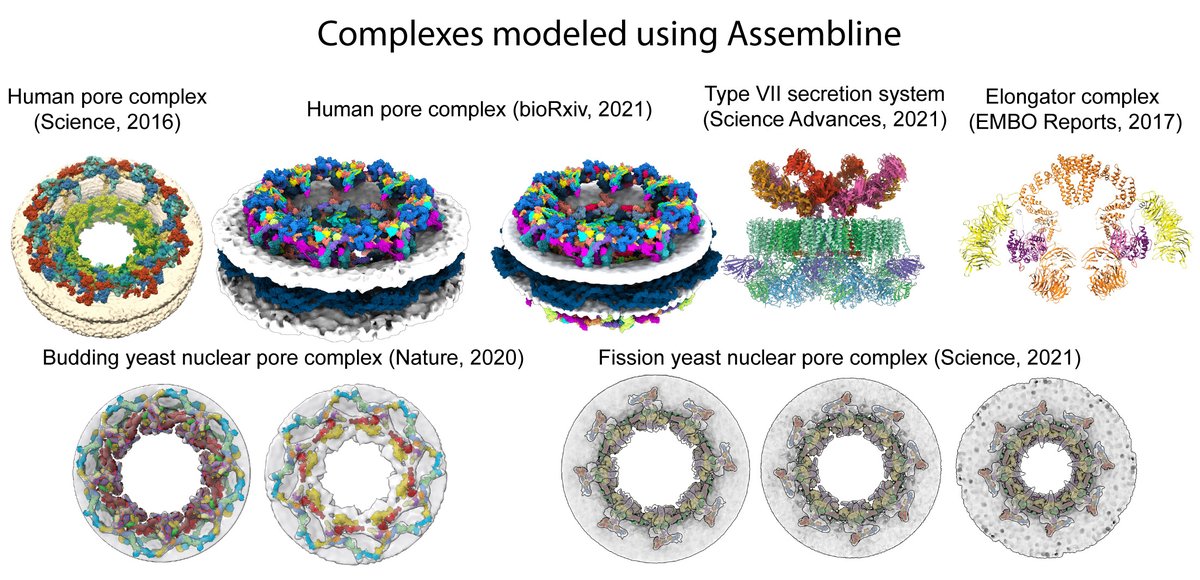
Our Assembline software, which we used for modeling nuclear pore and other complexes, is published today in Nature Protocols! Paper: nature.com/articles/s4159… Website: embl-hamburg.de/Assembline/

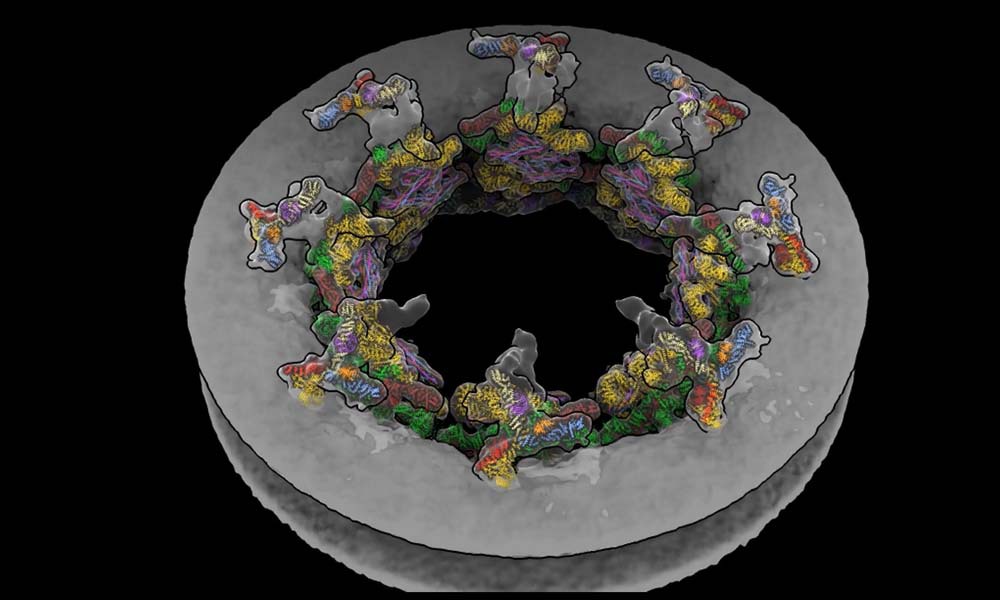
Watching molecules in the cell at atomic resolution: EMBL Hamburg’s Kosinski Group, Beck Laboratory & colleagues have visualised how the nuclear pore complex dilates and contracts in living cells, using a newly developed software. embl.org/news/science/o…


Our analysis of translation states in Dictyostelium is now online rdcu.be/c0RCb Subtomogram averaging from FIB lamellae at decent resolution by @pc_hoffmann Jan Philipp Kreysing Iskander Khusainov Sonja Welsch and Maarten Tuijtel. #CryoEM #TeamTomo Max Planck Institute of Biophysics


We listened to the community and here is the result: Got a CrossBeam that you would like to use for automated milling? The driver for lamella workflows in SerialFIB (github.com/sklumpe/Serial…), developed in collaboration with ZEISS Microscopy , has now been tested on 2 Crossbeams

Glad to finally share our manuscript on “Serial Lift-Out” by the amazing twitterless Oda Schiøtz, Jürgen Plitzko, Christoph Kaiser , and myself. Doing multiple sections from single lift outs showing beautiful C. elegans ultrastructure 🤯 I could watch it all day 🤩How did we get there?
It's out! It's out! 📢The bacterial Tc Toxins are out😱☠️ (and our paper 📜😉)! But how did these huge complexes get out? Where did they assemble? 🧐 Check out Tc Toxins' great escape from Yersina 🦠 Nature Microbiology: nature.com/articles/s4156… #cryoET #toxins #microbiology 🧵1/4
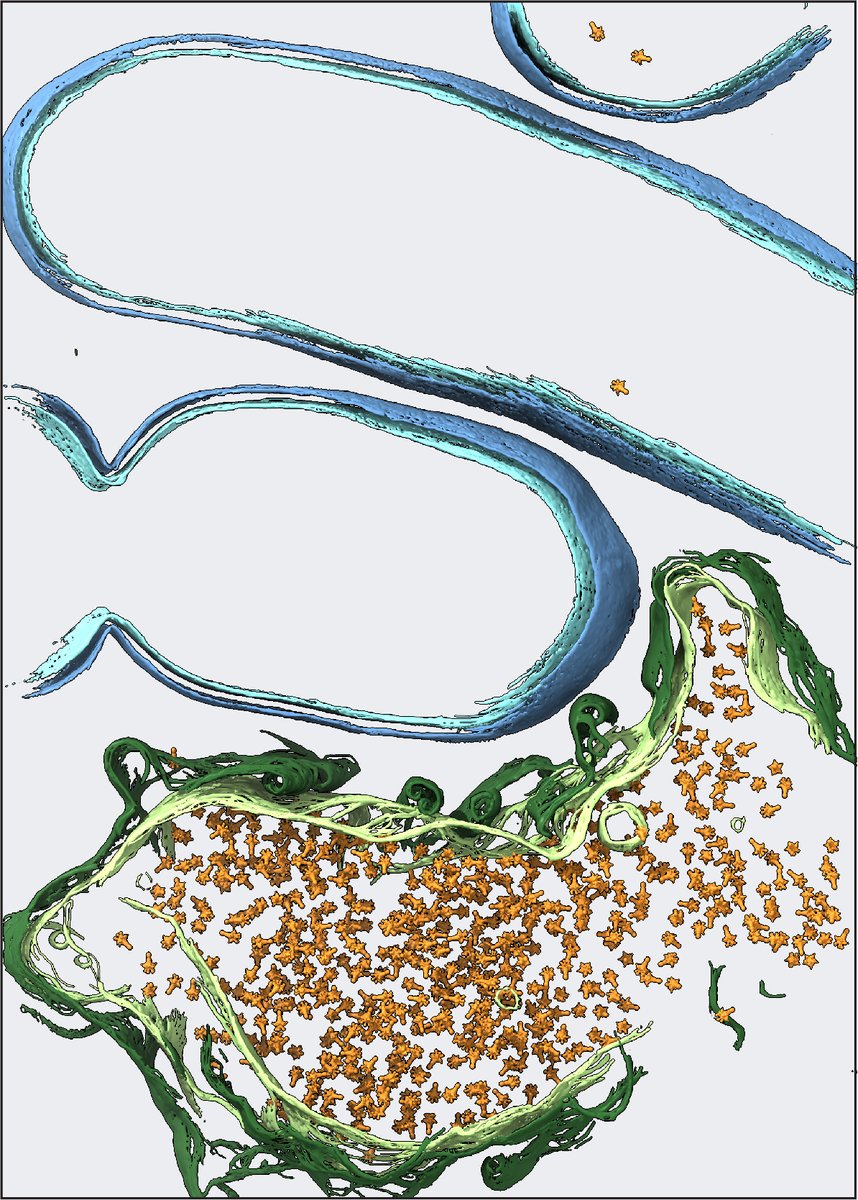



For your enjoyment: a mystery about replisome organization solved with twists and turns by Chen ZHANG (张琛). Why does the replisome sometimes appear as one bright and one dim vs two bright vs just one spot? Chen even discovered ways to pop the replisomes apart! rdcu.be/dFCod



Yeah congrats Jenny Sachweh and Bernhard and the whole Beck Laboratory crew! Llways something new with tose NPCs! Simply beautiful 😍