Marvin Albert
@_malberto
Bioimage analysis and computing at Institut Pasteur.
Previously at EMBL Heidelberg and UZH Zurich.
ID: 2277462703
05-01-2014 10:49:58
298 Tweet
437 Followers
1,1K Following

Let's go for Day 2 of the eRImote Euro-BioImaging ERIC distributed python course! Today's lesson on image segmentation and feature extraction is by France-BioImaging Research Infrastructure with Marvin Albert and Marie Anselmet 💻 Hosted in Lisbon by the GIMM Foundation Oeiras node.


🎉 Huge success at the "Intro to BioImage Analysis with Python for Life Scientists" course! Thanks to Euro-BioImaging ERIC, eRImote, Jean-Yves Tinevez, Rafael Camacho, and @gabymartins for making it happen! Inspiring to see #BioImageAnalysis community thriving! #AI4Life #PPBI #Python



1. Marvin Albert Marvin Albert will present *multi-view stitcher*, a Python framework for registration, stitching and fusion of multi-tiles, multi-view images.


2. Marie Anselmet could not join due to admin reasons, but Stephane Rigaud kindly volunteered to put her poster> In it we relate our WIP towards #TrackMate v8. This is a forthcoming new major version geared towards applications in microbiology.


3. Gaëlle Letort @gaLLetort has a poster showcasing two tools for developmental biology. The first one is #DeXtrusion, an AI tool built for the automated detection of rare events in time-lapse. Published here journals.biologists.com/dev/article/15… code here gitlab.pasteur.fr/gletort/dextru…


@gaLLetort 4. Minh-Son Phan Minh-Son Phan will present his recent work just published on the cell fate specification via lateral inhibition in Schweisguth’s lab of the @DevStem_Pasteur He spoke about it last week here: x.com/MinSonPhan/sta… .

@gaLLetort Minh-Son Phan @DevStem_Pasteur 5. Stéphane Rigaud Stephane Rigaud and Robert Haase will animate a workshop on #clEsperanto. CLE is a GPU-accelerated image processing library, that aims at offering lightning fast image processing with all GPU on all major OSes,

If you didn't yet have enough good reasons to share your bioimage data, Matthew (BioImage Archive), Josh (Josh Moore), Erin (@tuesdaystitches), Assaf (Assaf Zaritsky) and I are here with squirrel drawings to convince you that you should do it: journals.biologists.com/jcs/article/13…


We BIIF_Sweden had great fun teaming up with Frederic Ballllosera Navarro from Emma Lundberg's lab for an #i2k workshop! We discussed tools and approaches for #multiplex image data, and showed #PIPEX #TissUUmaps #spatialdata!


OME-Zarr: "Even a talk on formats [can be] interesting" Josh Moore Pavel Tomancak #I2K2024 #BioImageAnalysis
![Simon F. Nørrelykke (@simonflyvbjerg) on Twitter photo OME-Zarr: "Even a talk on formats [can be] interesting"
<a href="/notjustmoore/">Josh Moore</a> <a href="/PavelTomancak/">Pavel Tomancak</a> #I2K2024 #BioImageAnalysis OME-Zarr: "Even a talk on formats [can be] interesting"
<a href="/notjustmoore/">Josh Moore</a> <a href="/PavelTomancak/">Pavel Tomancak</a> #I2K2024 #BioImageAnalysis](https://pbs.twimg.com/media/Gaujug-WQAAmf51.jpg)


Amazing, Stephane Rigaud is demonstrating multi-GPU support🎉 using #pyclesperanto(without _prototype) for #BioImageAnalysis at #I2K2024(virtual)!🔬🖥️🚀 The notebook is available online #openaccess github.com/StRigaud/clesp…
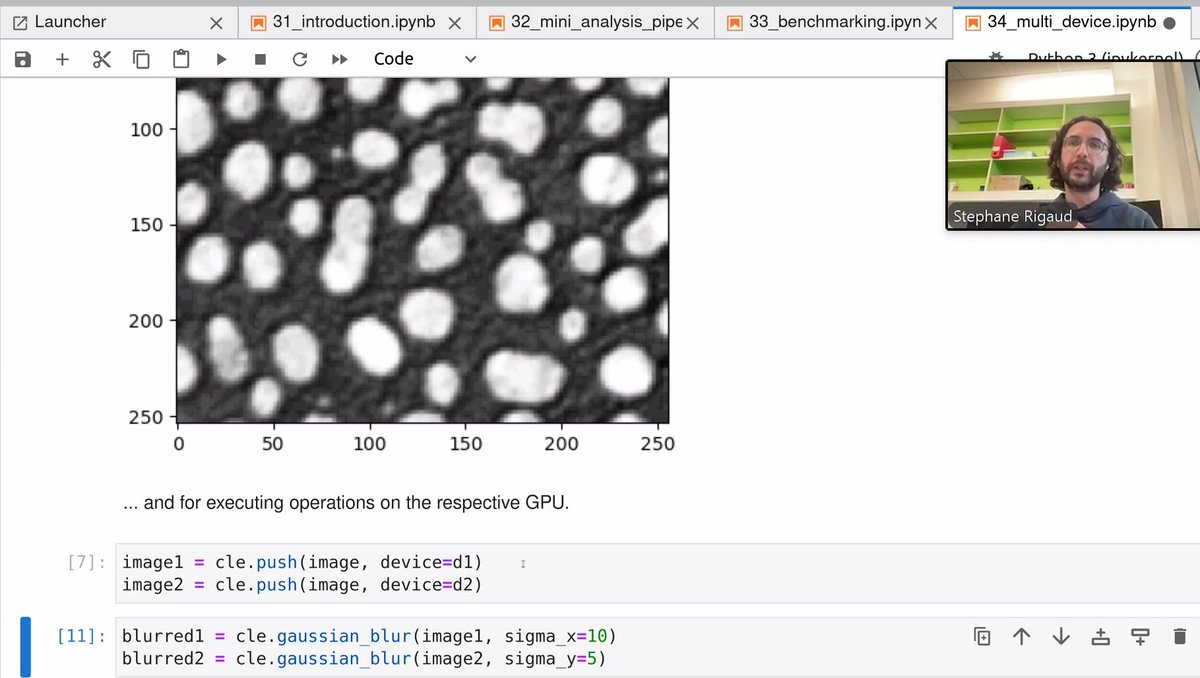


I'm presenting a poster at CZI Science Imaging The Future on the Three Gears of BioScience Projects. I will discuss how #imaging, analysis and modelling synergize and are fueled by clearly defined questions, and also how far should support go. 10.6084/m9.figshare.27725865