Nathanael Rollins
@_nathanrollins
learning from evolution + synthesizing biology scholar.google.com/citations?user…
ID: 800852856312131584
22-11-2016 00:06:14
74 Tweet
172 Followers
231 Following


Natural seq models are good at predicting pathogenic mutants- often better than wet-lab assays! Jonathan Frazer Mafalda Dias Pascal Notin Debora Marks classify 72,000 variants of unknown significance across human genome (& in total, 4x that in ClinVar!) biorxiv.org/content/10.110…

1/n I'm excited to share our preprint *Large-scale clinical interpretation of genetic variants using evolutionary data and deep learning* biorxiv.org/content/10.110… from a great collab with Pascal Notin Mafalda Dias @AidanNGomez Dr. Kelly Brock Yarin (OATML_Oxford) Debora Marks 🧵


Can one anticipate viral evolution enough to support timely intervention? Check out our new preprint on forecasting antibody escape biorxiv.org/content/10.110… With Sarah Gurev Pascal Notin Noor Youssef Nathanael Rollins Chris Sander aka cscbio Yarin (OATML_Oxford), Debora Marks, 🧵1/10


We’ve updated our EVEscape paper with more SARS2 retrospective evaluation and potential use-cases! biorxiv.org/content/10.110… Collaborators Sarah Gurev Pascal Notin Noor Youssef Nathanael Rollins Chris Sander aka cscbio Yarin (OATML_Oxford), Debora Marks, 🧵1/4



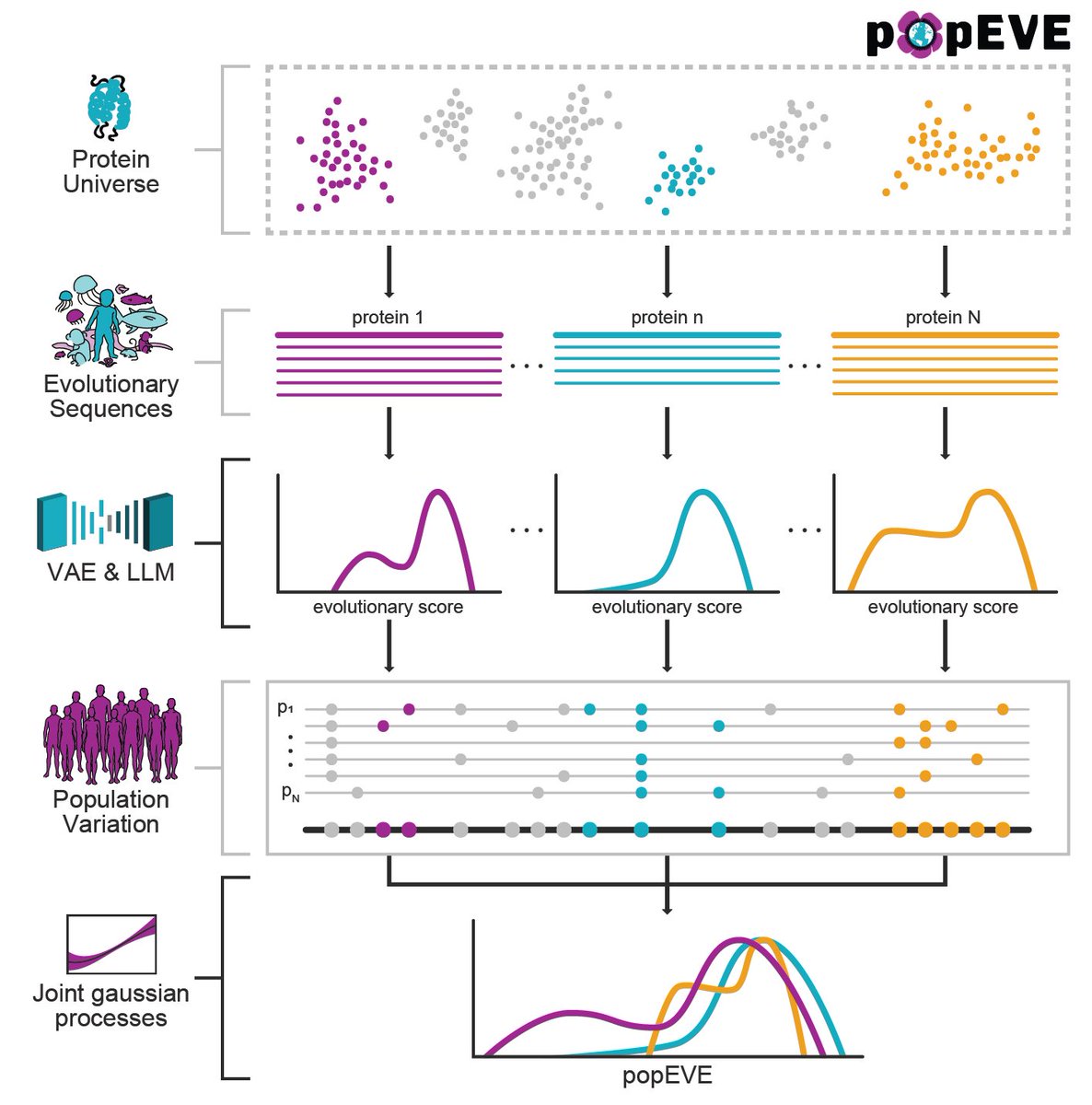
Announcing popEVE - a deep generative model of the human proteome that reveal over a hundred novel genes involved in rare genetic disorders medrxiv.org/cgi/content/sh… Jonathan Frazer Aaron Kollasch @H__Spinner Courtney Shearer Mafalda Dias Debora Marks explore at pop.evemodel.org