Alessandro Lussana 🇺🇦
@a_lussana
alussana.xyz
ID: 1725937591
http://alussana.xyz/ 03-09-2013 15:52:39
151 Tweet
84 Followers
190 Following

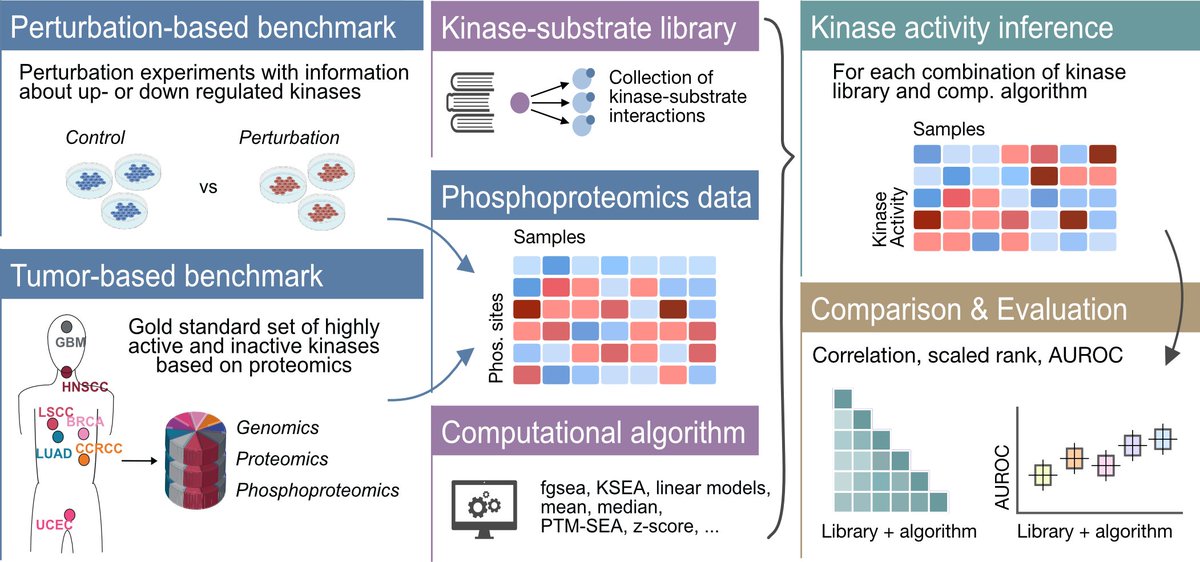
Ever wondered which method to use to infer kinase activities from phosphoproteomics data? 💻💭In collaboration with the Zhang lab BCMHouston, we performed a comprehensive evaluation of kinase activity inference tools to help answer that question tinyurl.com/yc56axsv 🧵⬇️

Introducing *fullRankMatrix*, available on CRAN! Constantin Ahlmann-Eltze (@const-ae.bsky.social) and I developed this R package to construct full rank design matrices. Esp. relevant for binary matrices that contain linearly dependent columns. Use fullRankMatrix for accurate linear model interpretation! 📊 #rstats



Our postdoc Iguaracy Pinheiro-de-Sousa Iguaracy Pinheiro-de-Sousa is using multiomics data to help develop better organoids and to understand how the heart🫀 and brain 🧠work at a molecular level. Collaboration and knowledge exchange are key in Iguaracy's work. ebi.ac.uk/about/news/per…


Matthias Matthias Mann Lab shows first joint proteome and RNA workflow in same cell - very exciting, and lots of promises to build multimodal atlases in nearer future!




Wonderful time presenting at UW–Madison with Federico Marotta. Meeting talented scientists dedicated to driving #compbio towards a positive impact on society was truly inspiring. Many thanks to ISCB News for inviting us as the winning team of the PEGS DREAM Challenge to this great event!


PhosX is out! doi.org/10.1093/bioinf… Leveraging the latest kinase specificity maps from the recent monumental papers from the Cantley Lab Michael Yaffe Turk Lab and a rank sum statistic we created PhosX so now you can predict activities for near all kinases in your phosphodata :)
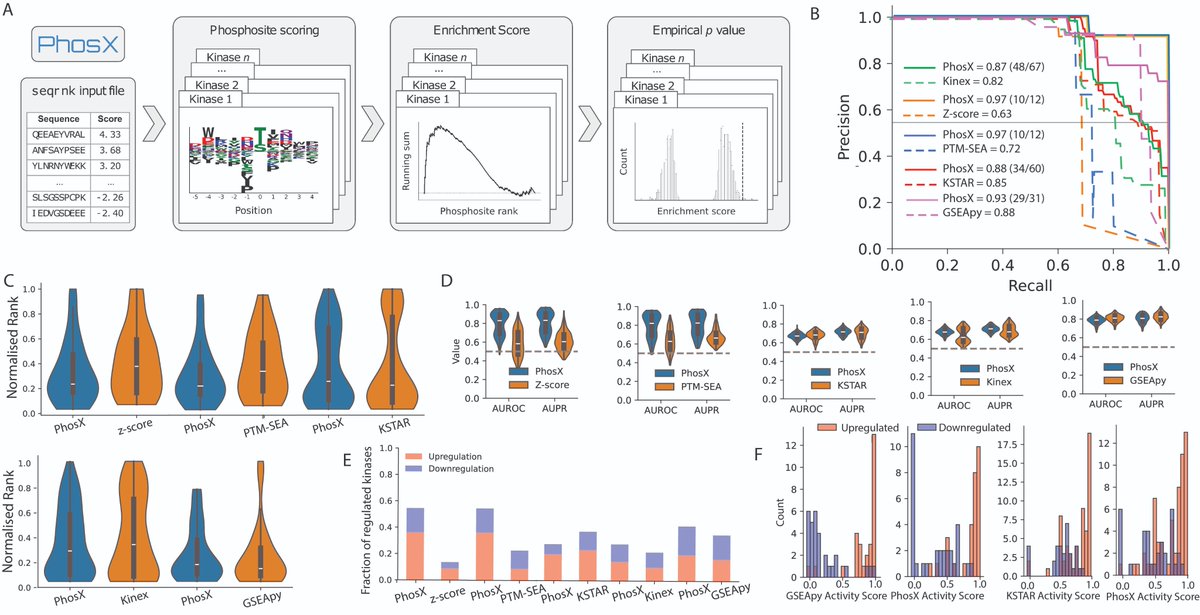

Kinases play a crucial role in many diseases, making them prime targets for therapeutic intervention. Enter PhosX, a new tool transforming the way researchers analyse and interpret kinase activity. Read the full article👇: academic.oup.com/bioinformatics… Alessandro Lussana 🇺🇦 Evangelia Petsalaki



Ever wondered which method to use to infer kinase activities from phosphoproteomics data? 💻💭Our revised comprehensive evaluation of kinase activity inference tools, done in collaboration with the Zhang lab BCMHouston, is now out Nature Communications 🔬 tinyurl.com/4twuc6z4