Abhishek Singharoy
@abhisekhsingha1
Biophysicist, Integrative modeler, Prof @ASU, Director of DoD's biosense.mn.co, #nsfcareer awardee, Beckman Fellow, Top Performer @NSF Waterman Fellow
ID: 1034465541749178375
https://search.asu.edu/profile/3146260 28-08-2018 15:39:39
541 Tweet
901 Followers
191 Following

Great turnout at yesterday’s BPS Arizona Student Chapter Science Trivia! 🧠✨ Gift cards will be sent out soon to our winners. Thanks to everyone who participated and made the event a success. Here’s to more fun and learning in science! 🔬💡



The BPS Arizona Student Chapter Biophysical Society activities make me a really proud mentor !! Calendars coming soon :)

#JobOpening The group is looking for *2 new graduate students* in Computational Biophysics (QM/MM; MD; informatics, systems biology broadly defined) for start in the Fall, 2025. Plz apply to ASU School of Molecular Sciences, and prompt us in your app. RT & mail me app directly !

Wishing a Happy 2025 from the entire team ASU School of Molecular Sciences & Biodesign Institute at Arizona State University !! Sounds on for the background music. It synced remarkably well this time :)

First paper of 2025 with Ross Wang Lab & #AlisonOndrus Here is a cute and practical technique by Sumit for studying reactions under external force, complementing #CoGEF methods. #compchem cell.com/heliyon/fullte… Heliyon ASU School of Molecular Sciences Chemistry at UIC






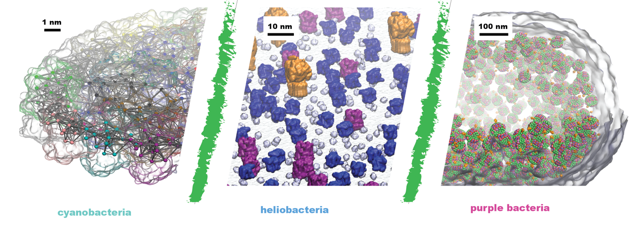
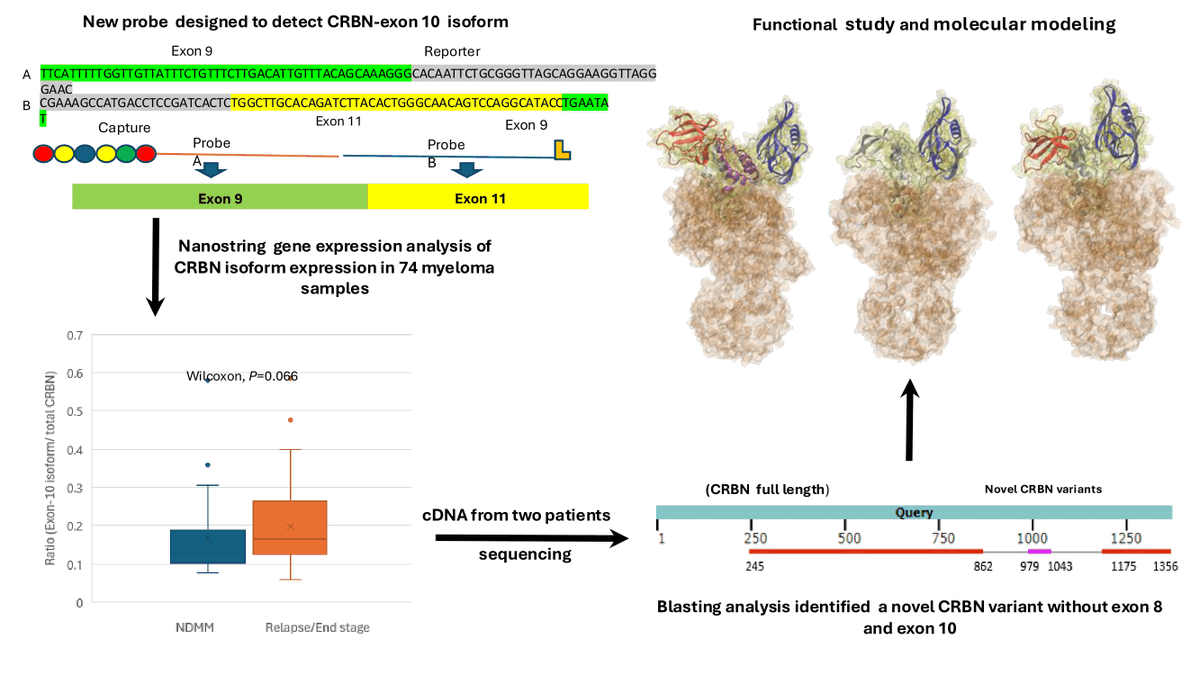
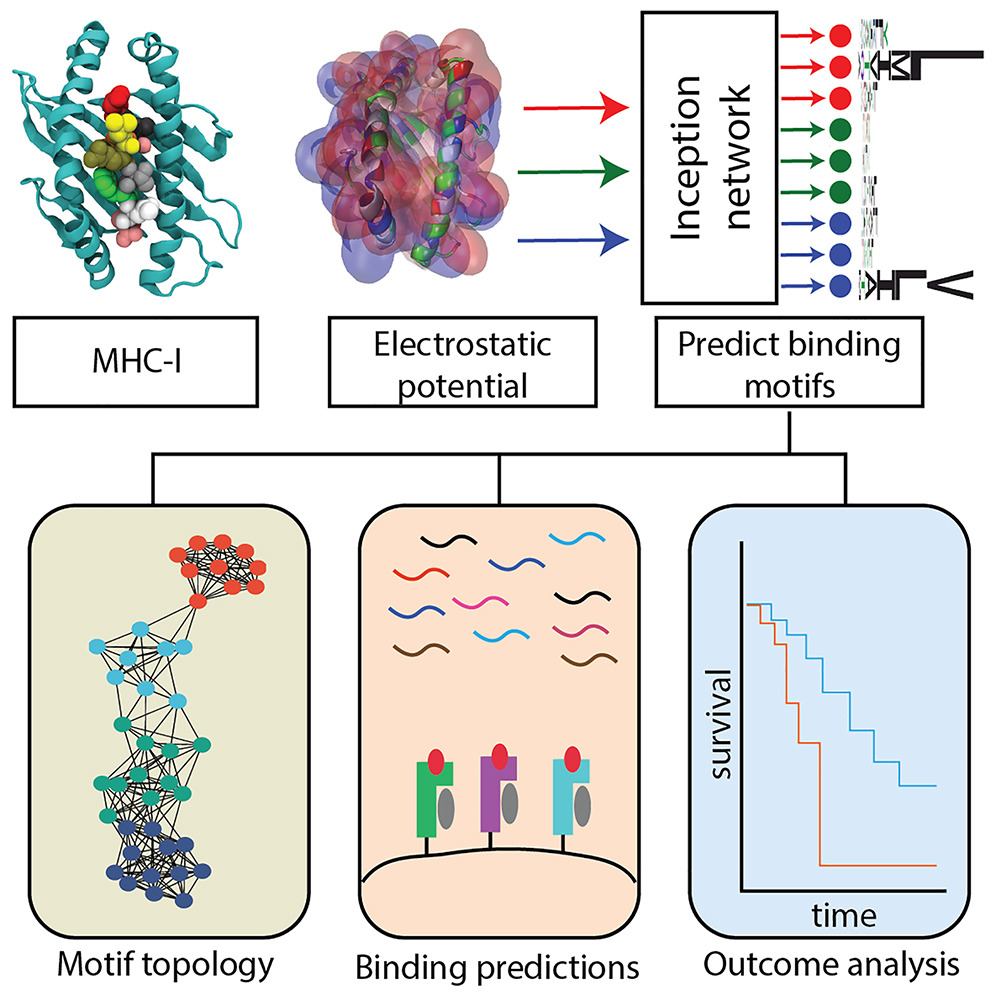
Amazing images we received this year😁 Discover! Disseminate! Develop! Biophysical Society