Anand Divakaran
@adivkrn
🇮🇳🇺🇸👨🔬 Applying medicinal chemistry & chemical biology to study epigenetics @DanNomura lab.
🎓PhD @UMN_MedChem.
🦮 Former CanDoCanines puppy raiser
ID: 832267612264468481
16-02-2017 16:37:16
244 Tweet
238 Followers
313 Following

New Smith Lab from Medical College of Wisconsin of WI publication in ACS Publications Bio & Med Chem Content Journal of Medicinal Chemistry: "Selective and Cell-Active PBRM1 Bromodomain Inhibitors Discovered through NMR Fragment Screening" doi.org/10.1021/acs.jm… #bromodomains #NMR #PBRM1 #SAR

Nice paper by Kevan Shokat using ligand-observed 19F NMR for probing differential binding to KRas. A fun read. pubs.acs.org/doi/10.1021/ac…

Excited to present our bioRxiv on chemical rational design of molecular glue degraders through a covalent handle targeting RNF126! Congrats to Ethan Toriki & James Papatzimas who led work w/Novartis Science! & thanks to The Mark Foundation for Cancer Research ! biorxiv.org/content/10.110…

I'm so excited to have this work coming out on our rational design of molecular glue degraders! We converted a number of inhibitors into glues with a simple covalent handle. Such a great project working alongside my good buddy Ethan Toriki and under the guidance of Dan Nomura!


Doing our due diligence celebrating Ethan Toriki and James Papatzimas getting this heroic piece of work out, great effort guys!


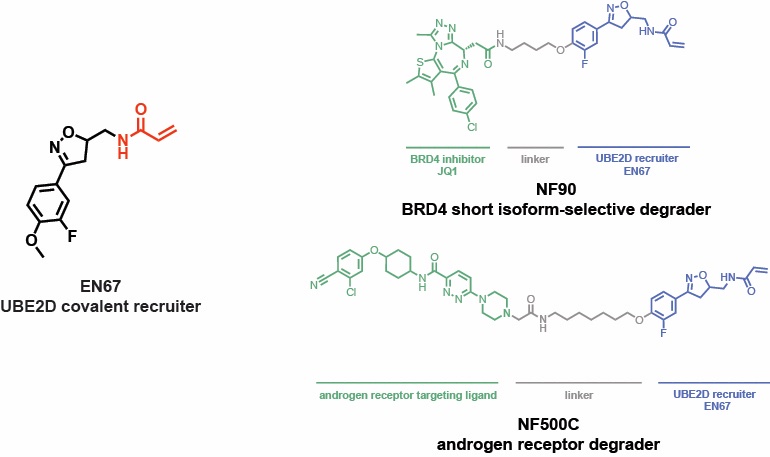
Excited to showcase our latest work by postdoc Nafsika Forte College of Chemistry UC Berkeley MCB & Novartis Science on covalently recruiting E2 ubiquitin conjugating enzyme UBE2D for targeted protein degradation applications!! Also thanks to The Mark Foundation for Cancer Research! biorxiv.org/content/10.110…

Proud to see Frontier Medicines announce FMC-376, a covalent inhibitor that directly, rapidly, and completely blocks both active and inactive forms of KRAS G12C to overcome non-response and resistance seen with prior generation KRAS G12C inhibitors! Yay!! globenewswire.com/news-release/2…

Grateful to see our work highlighted alongside leaders in the field @craigmcrews Ingrid Wertz The Amit Choudhary Lab Stuart Schreiber in the exciting and exploding area of proximity-inducing modalities for drug discovery in Nature Reviews Drug Discovery nature.com/articles/d4157…

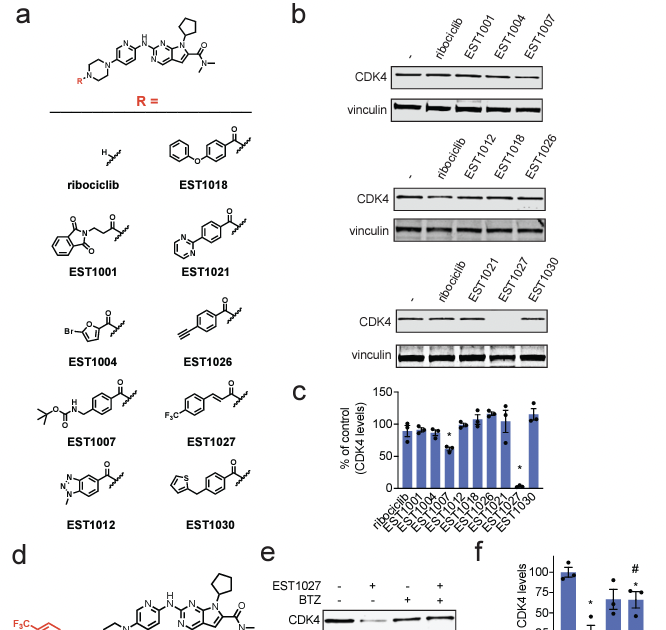
Congrats to Nafsika Forte for the publication of her paper in ACS Chemical Biology on the discovery of a covalent E2 ubiquitin conjugating enzyme UBE2D recruiter for PROTAC applications! Thanks to Novartis Science. And thanks to The Mark Foundation for Cancer Research ! pubs.acs.org/doi/10.1021/ac…



Back and recovered from a stimulating Bioorganic GRC Taking back a lot of new ideas and wonderful memories from catching up with lab alumni, old friends and new. with Anand Divakaran @ColeScholtz Nora Vail Ross Cheloha DykhuizenLab Thanks Dan Nomura!



Excited to showcase our new bioRxiv frm postdocs @floraliciaflor & Nafsika Forte on the discovery of covalent degraders of oncogenic transcription factor b-catenin (CTNNB1) via destabilization! College of Chemistry UC Berkeley MCB The Mark Foundation for Cancer Research biorxiv.org/content/10.110…


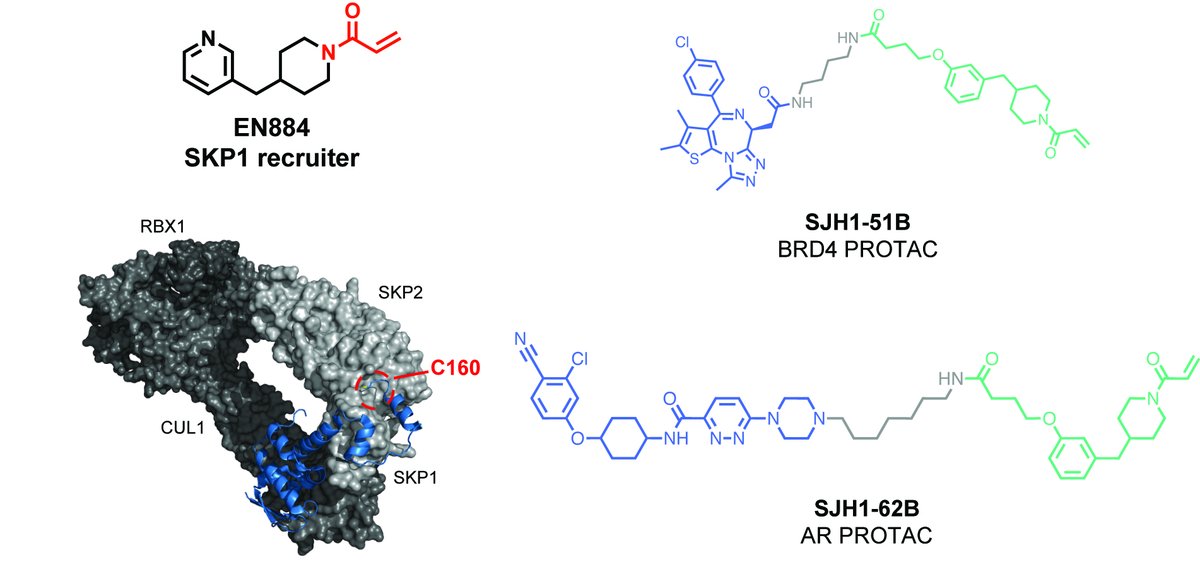
Congrats to our postdocs Seong Ho (Johnny) Hong & Anand Divakaran on the discovery of a covalent recruiter against the CUL1 substrate adaptor protein SKP1 that targets a SKP1/substrate receptor interfacing C160 to enable PROTAC applications! College of Chemistry UC Berkeley MCB The Mark Foundation for Cancer Research

Congrats to Melissa Lim & Thang Do Cong on their ACS Central Science paper on the discovery of a minimal degradative covalent handle that targets DCAF16 for rational design of monovalent degraders! Thanks Novartis Science The Mark Foundation for Cancer Research! College of Chemistry UC Berkeley MCB pubs.acs.org/doi/10.1021/ac…