
Danny Diaz
@aiproteins
Changing the World! One protein at a time! ML Protein Engineer | DeepProteins @MLFoundations | Entrepreneur
ID: 1005975495400411138
http://danny305.github.io 11-06-2018 00:50:23
644 Tweet
407 Followers
418 Following


Of >105,000 participants with 30-year follow-up, only 9.3% achieved healthy aging (age 70, w/o any chronic diseases). Their diet was significantly associated with this outcome🧵 Nature Medicine


Small proteins can be more complex than they look! We know proteins fluctuate between different conformations- but how much? How does it vary protein to protein? Can highly stable domains have low stability segments? Állan Ferrari experimentally tested >5,000 domains to find out!


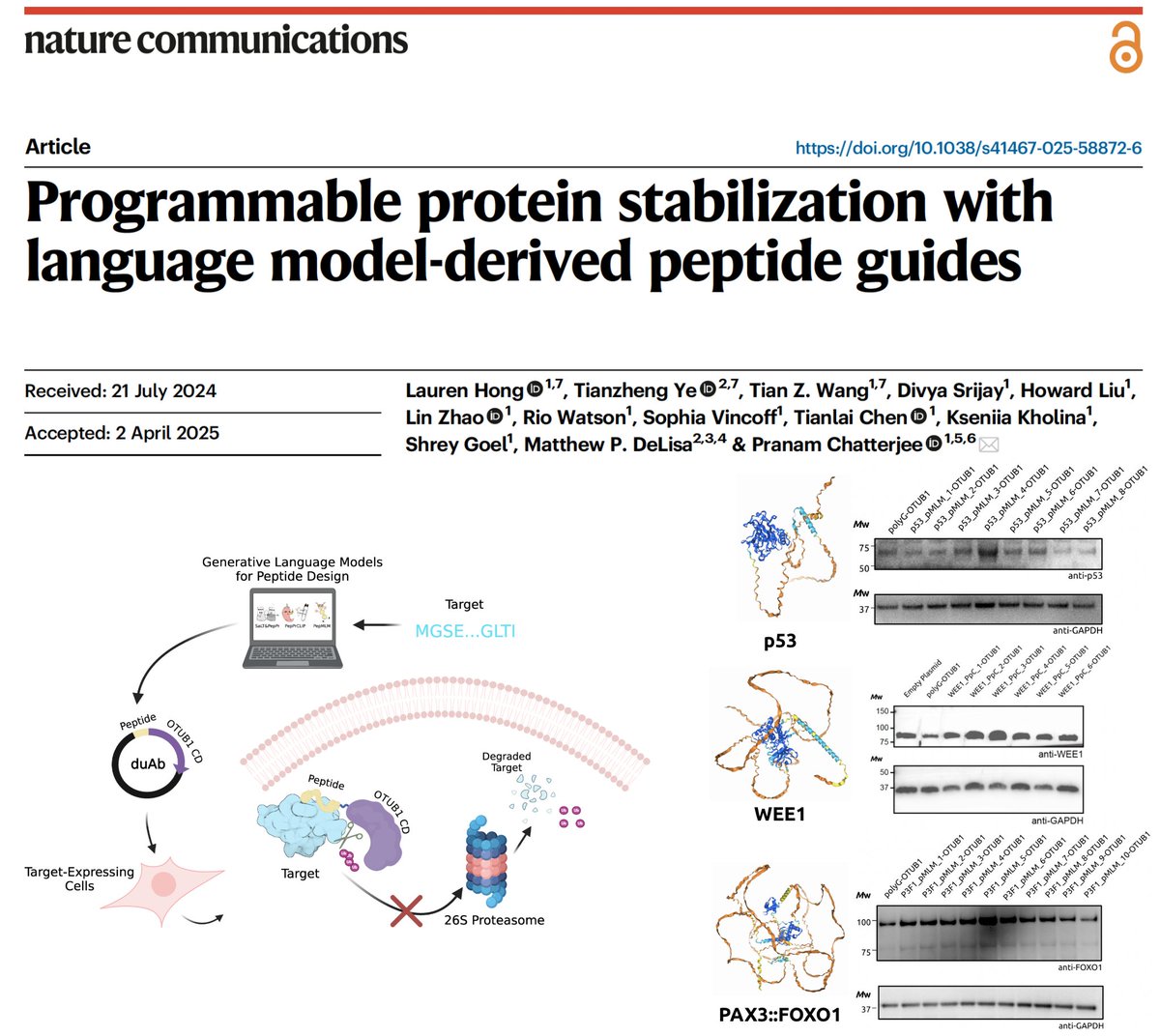
Our pLM-designed peptides, when attached to E3 ligases (aka uAbs), enable us to programmably degrade disease targets (think CRISPRi, but for proteins). But what if we want to STABILIZE useful proteins? In Nature Communications, we introduce duAbs! 🌟 📜: nature.com/articles/s4146… 🧬:








Gaia: An AI-enabled genomic context–aware platform for protein sequence annotation Science Advances 1.Gaia introduces a new paradigm in protein function annotation by combining sequence, structural, and crucially, genomic context information into a unified, scalable search





Had a lot of fun learning diffusion and addressing key issues in protein diffusion with Giannis Daras Jeffrey Ouyang-Zhang TLDR: a few protein structure insights inspired us to design a new diffusion loss, training regime and dataset, resulting in significant performance improvements










