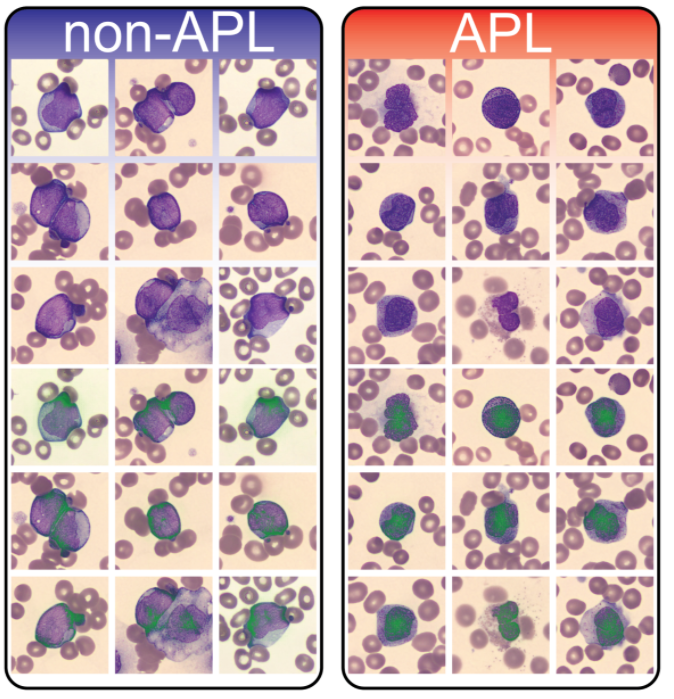
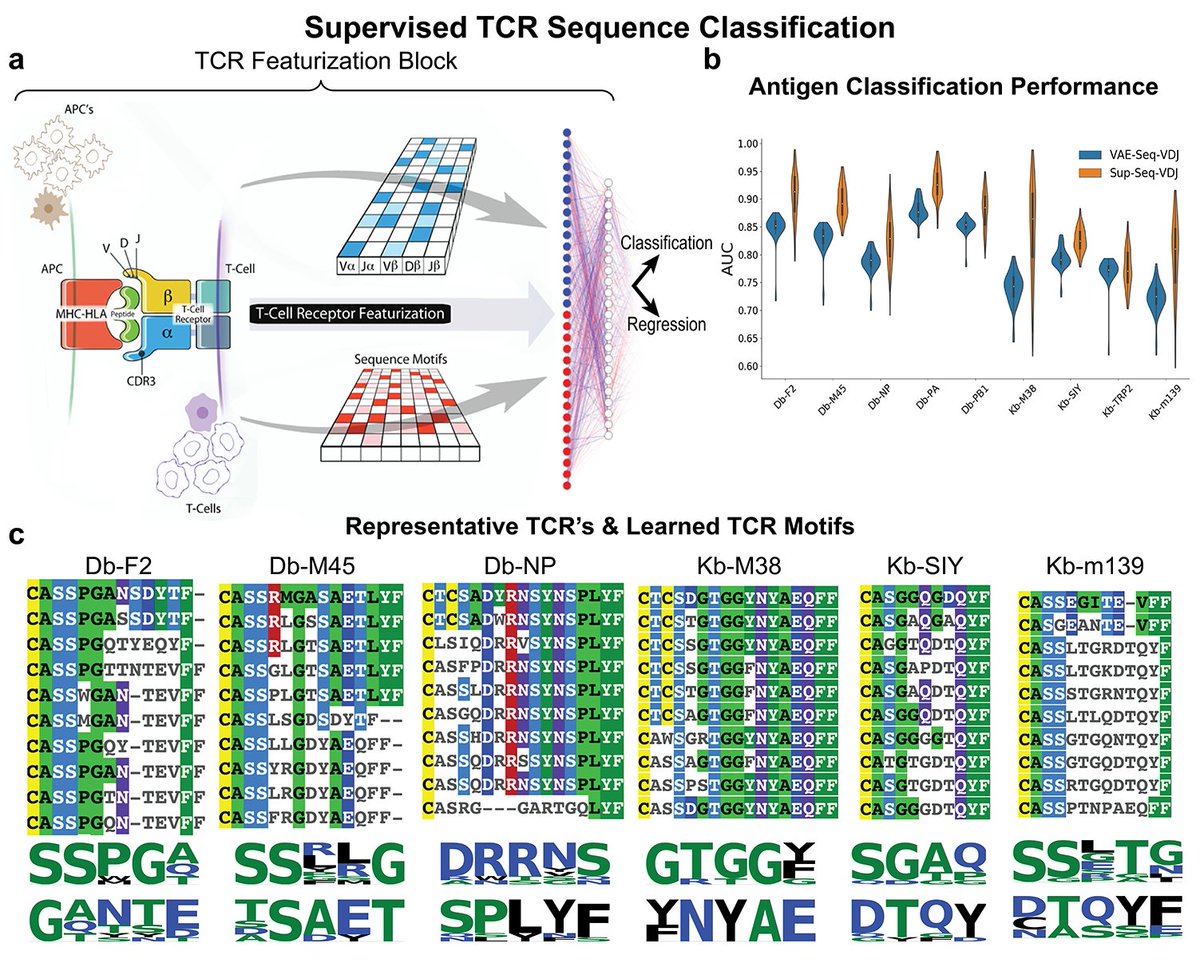
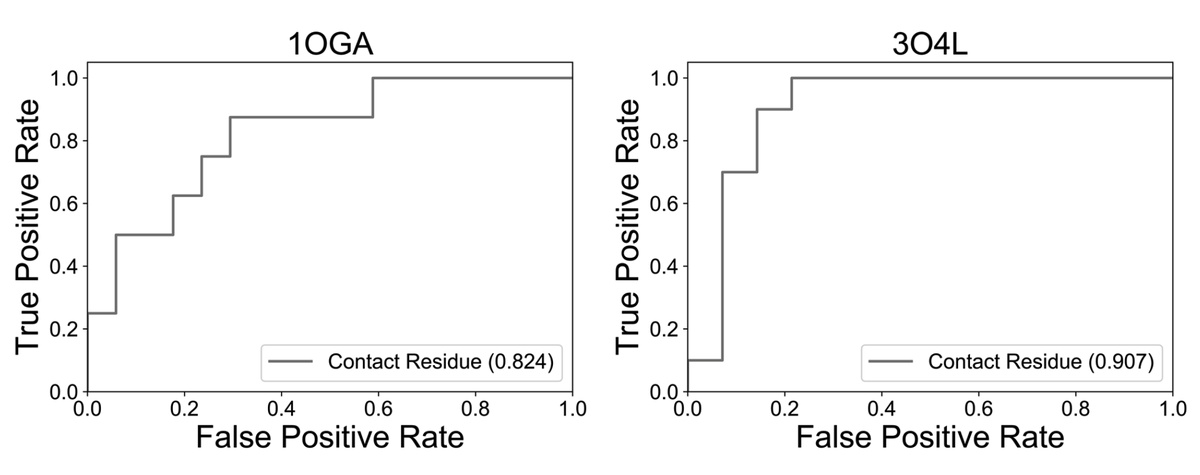
Alex Baras
@alexander_baras
Associate Professor of Pathology, Urology, and Oncology.
Director of Precision Medicine Informatics.
Johns Hopkins Sidney Kimmel Comprehensive Cancer Center.
ID: 1303037109683589120
07-09-2020 18:27:23
16 Tweet
61 Followers
18 Following