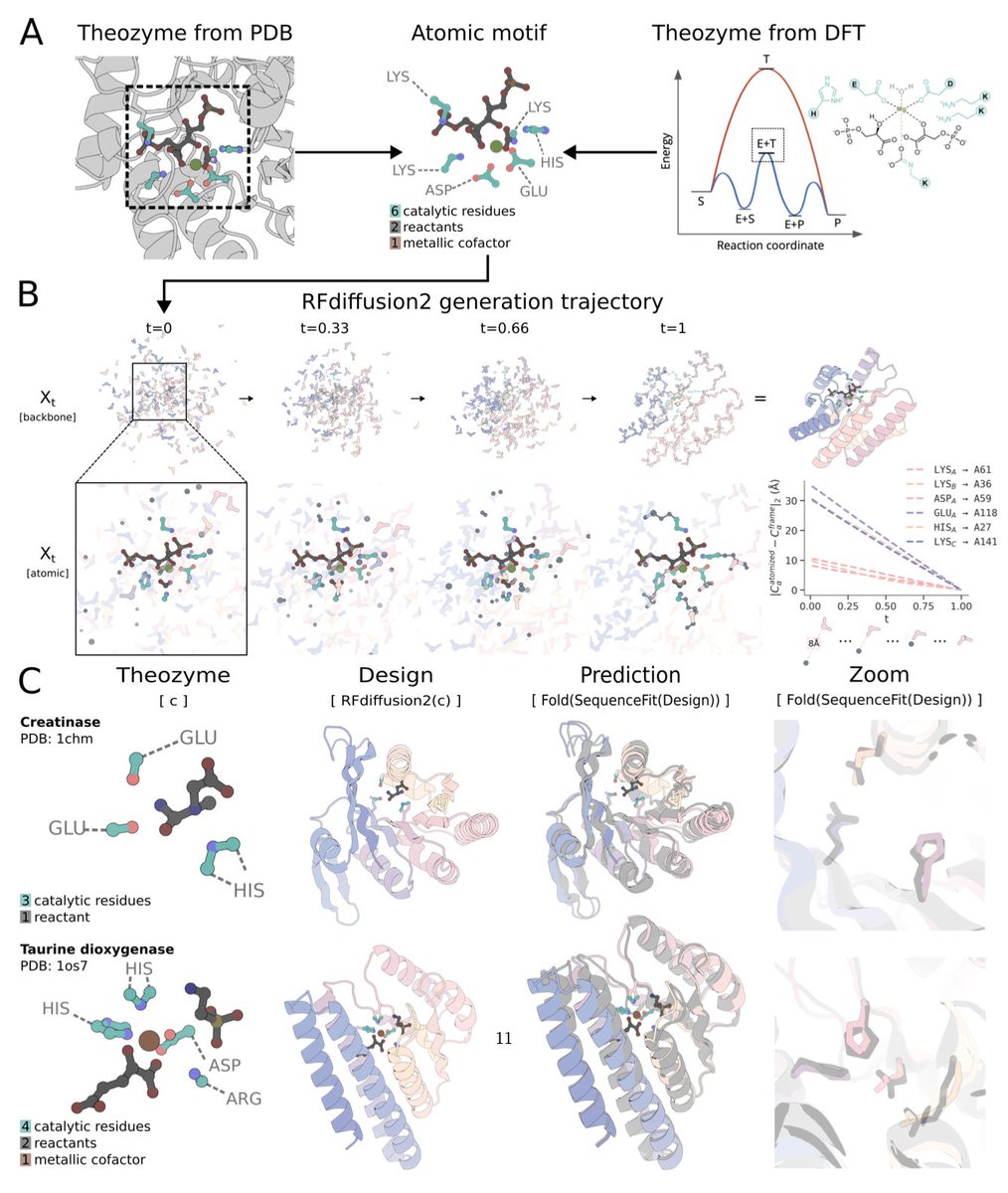
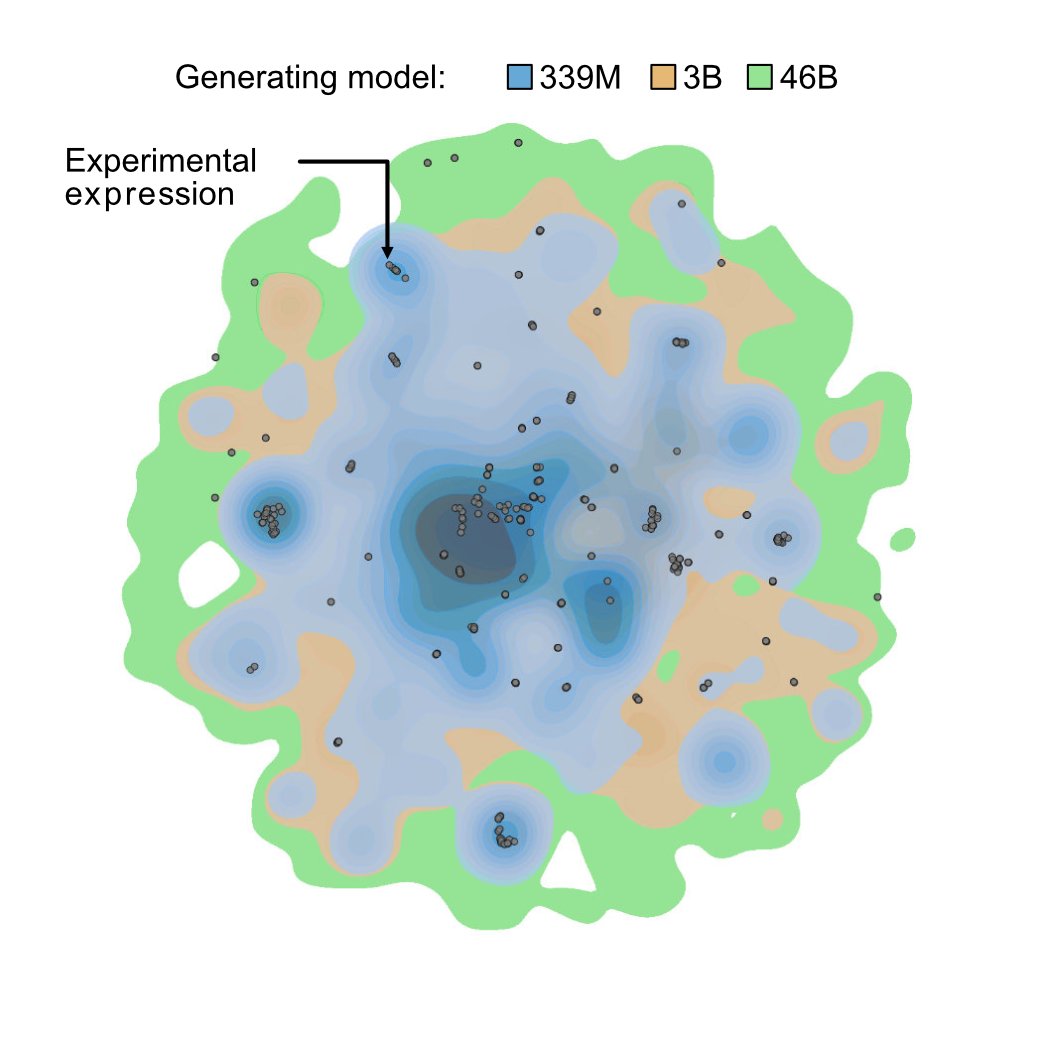
Alex Chu
@alexechu_
Deep learning & protein design. @googledeepmind PhD @Stanford
ID: 3013702976
http://alexechu.github.io 08-02-2015 19:08:36
667 Tweet
1,1K Followers
749 Following

Excited to share our preprint “BoltzDesign1: Inverting All-Atom Structure Prediction Model for Generalized Biomolecular Binder Design” — a collaboration with Martin Pacesa, Zhidian Zhang , Bruno E. Correia, and Sergey Ovchinnikov. 🧬 Code will be released in a couple weeks









📢 Excited to present our poster at the #ICLR2025 GEMBio Workshop! I'll be introducing TherAbDesign - a novel, sequence-based framework that efficiently optimizes antibodies toward therapeutically relevant biophysical properties. [1/4]
![Amy Wang (@amywang01) on Twitter photo 📢 Excited to present our poster at the #ICLR2025 <a href="/gembioworkshop/">GEMBio Workshop</a>! I'll be introducing TherAbDesign - a novel, sequence-based framework that efficiently optimizes antibodies toward therapeutically relevant biophysical properties. [1/4] 📢 Excited to present our poster at the #ICLR2025 <a href="/gembioworkshop/">GEMBio Workshop</a>! I'll be introducing TherAbDesign - a novel, sequence-based framework that efficiently optimizes antibodies toward therapeutically relevant biophysical properties. [1/4]](https://pbs.twimg.com/media/GpbLx1fbgAAqDaO.jpg)


Excited to share new validation results of our Google DeepMind Google AI AI co-scientist! Addressing the challenge of liver fibrosis, a condition affecting millions globally with few treatments options, we've been working with Dr. Gary Peltz Stanford Medicine, exploring how








I've been working on variants of this problem for my entire career, and have only dreamed of designing molecules on the computer with success rates this high. Proud of Chai Discovery for pulling this off, and doing it with the speed, rigor and grace that was only imaginable ✨