Martin Villalba
@amartinvill
ID: 3397097926
http://martin-villalba-lab.github.io/ 31-07-2015 12:22:35
119 Tweet
308 Followers
74 Following

Registration for the 24th Biennial Meeting of the International Society for Developmental Neuroscience is open! Submit abstracts by April 9 ,2022 #DevelopmentalNeuroscience ISDN 2024 Conference

Happy to share our recent paper published in Science Advances Science Advances The mechanisms of astrocyte increase during evolution, and its role in cortical folding. Localized astrogenesis regulates gyrification of the cerebral cortex science.org/doi/10.1126/sc…


Submitted my abstract for the #CSHLglia meeting 2022 to represent Martin-Villalba Lab Cold Spring Harbor Laboratory! So excited to meet the community and share/discuss the results of my PhD project 👨🏽🔬

After a couple of great years at Mainz University, I recently returned to DKFZ to team up again with Prof. Dr. Frank Lyko and the fantastic scientists of my beloved DKFZ Epigenetics. I am really very happy to be back! See you in Heidelberg from now on for more & better science🧬!



Congrats to the Vorholt Microbiology_ETH and @BartDeplancke for their landmark paper on Live-seq, a method for recording the transcriptome from single cells without killing them! ETH news article: ethz.ch/en/news-and-ev… Paper: rdcu.be/cTPLe



Here it is: our #inception paper Development „De novo PAM generation to reach initially inaccessible target sites for base editing“ by Kaisa Pakari & Thomas Thumberger We use 2 guides & 2 editors to edit distant nucleotides with NGG base-editors journals.biologists.com/dev/article-ab… #medaka #DNA

A huge thanks to everyone who made this study possible: my mentor Irv Weissman, the lab's brain team (Rahul Rahul Sinha, Nobuko, Joy, Anna et al.), as well as Chan Zuckerberg Biohub Network and Stanford MSTP for their support!


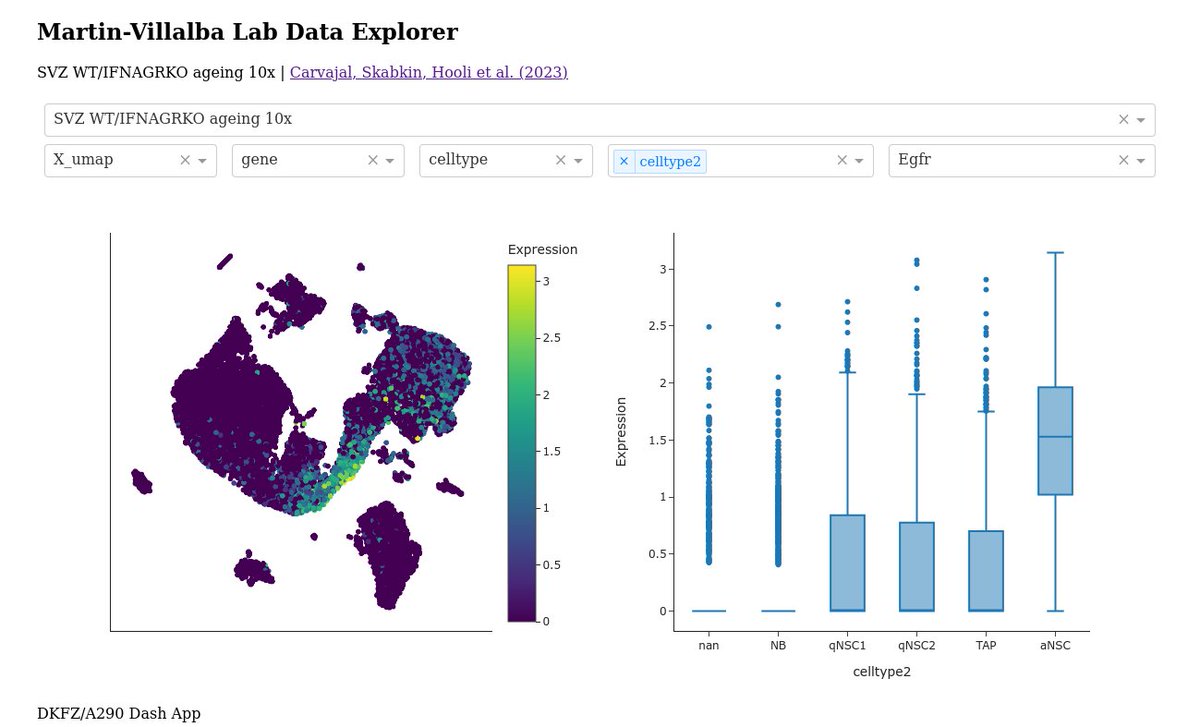
I have gathered up a bunch of published #SingleCell datasets from Martin-Villalba Lab and made this thing: a290-data.dkfz.de Nearly 27000 subventricular zone cells from three different publications available for exploration!





We're so excited to share our latest work “Identification of astrocyte-driven pseudolineages reveals clinical stratification and therapeutic targets in #Glioblastoma” spearheaded by Oğuzhan Kaya on the wet & @LCForst on the dry lab as a preprint @Biorxiv biorxiv.org/content/10.110…

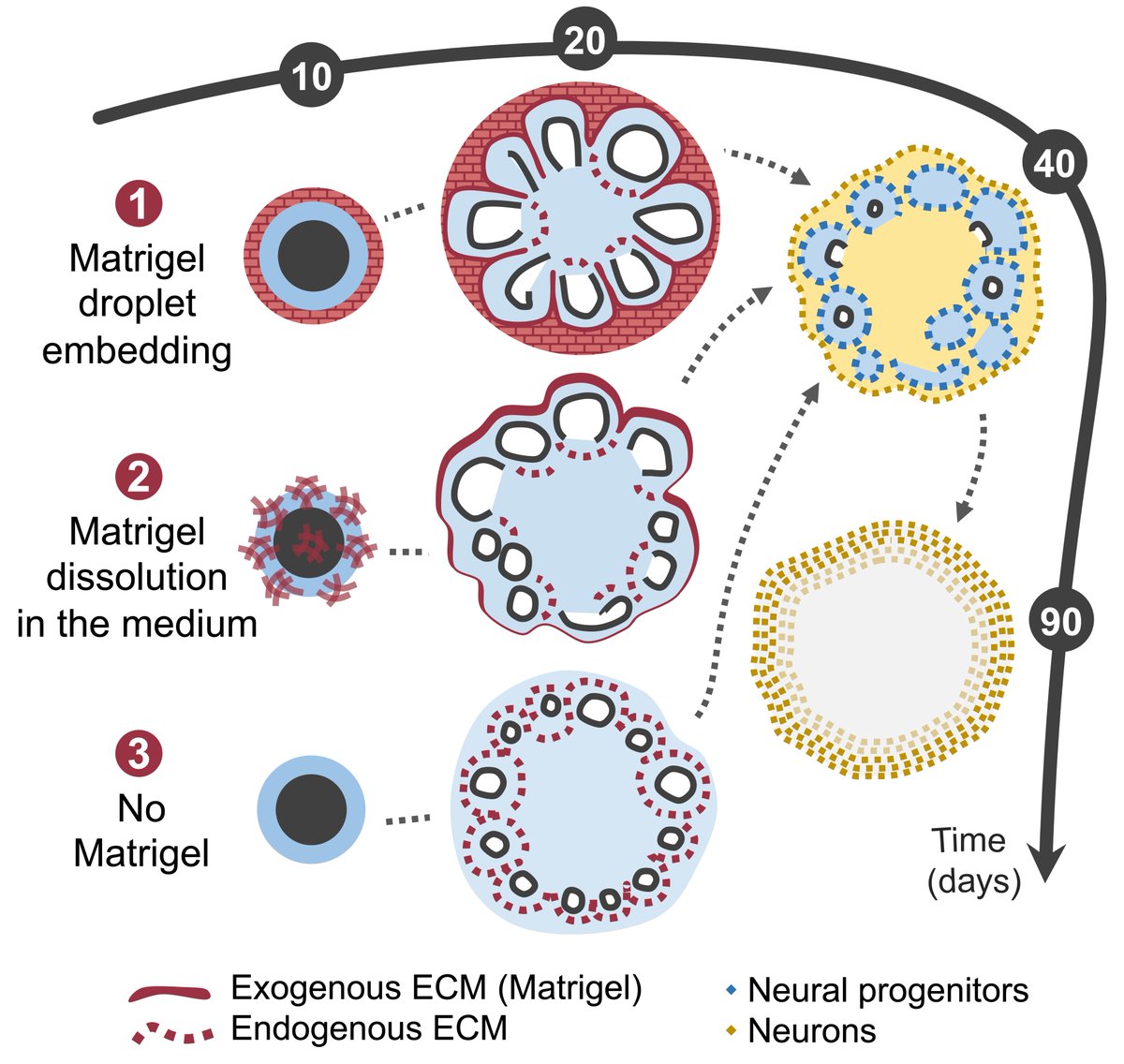
📢 We’re live at the The EMBO Journal! 🌐 tinyurl.com/39xdttna Human telencephalic organoids cultured in the absence of Matrigel endogenously produce and self-organize the extracellular matrix, without affecting programs of neuronal differentiation. Led by Catarina Martins Costa 👩🔬 🧵 1/5