
Ruibang Laurent Luo
@aquaskyline
Associate Professor of Computer Science at HKU. Private Pilot. #bioinformatics/#engineering.
ID: 109840044
http://luo-lab.hk 30-01-2010 13:10:52
826 Tweet
805 Followers
345 Following





Can deep learning improve variant calling accuracy in long-read RNA sequencing data despite its inherent challenges like high error rates and uneven coverage? The University of Hong Kong Rice University bioRxiv "Clair3-RNA: A deep learning-based small variant caller for 2 long-read
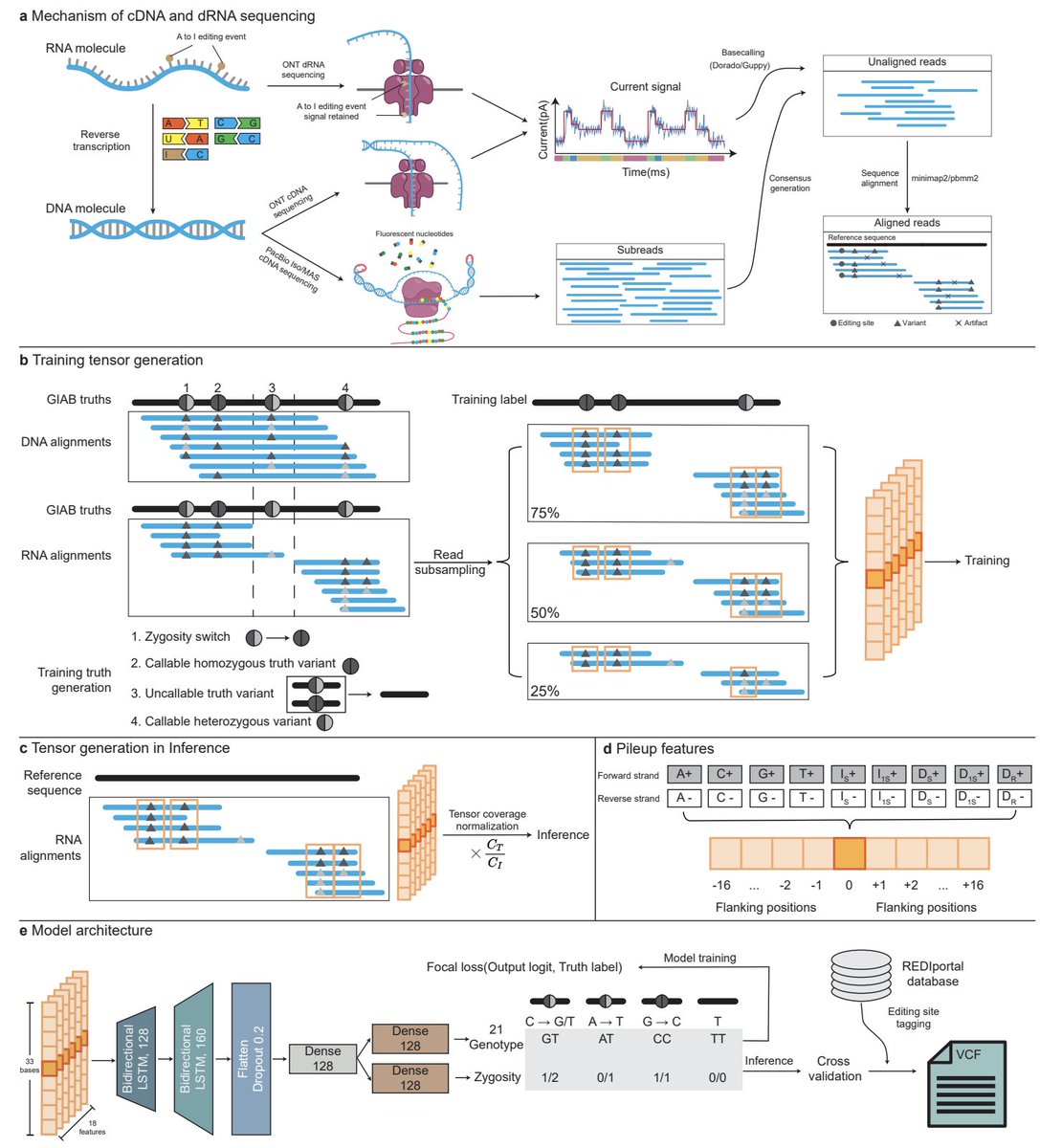

The preprint of Clair3-RNA is now in bioRxiv 🔗biorxiv.org/content/10.110…. Phasing gives F1 another 5% jump, and is now available in v0.2.0. Great collaborating again with Fritz Sedlazeck.

I had my hopes to call it ClaiRR to make a pirate theme tool but Ruibang Laurent Luo stayed professional... what can I say it all started with Margaritas in Houston :) haha









