
Min Dai
@artofbiology
Senior computational scientist in Fishell lab at the Broad Institute and HMS. Ph.D. and B.S. in Bioinformatics. Interests: #SingleCell #MachineLearning #Neuron
ID: 849983919587971072
https://genecell.github.io/ 06-04-2017 13:55:33
2,2K Tweet
1,1K Followers
950 Following




Two recent papers about cellular segmentation were published on Nature Methods Nature Methods : 1. Segment Anything for Microscopy nature.com/articles/s4159… 2. Cellpose3: one-click image restoration for improved cellular segmentation nature.com/articles/s4159…

Simultaneous analysis of single-cell gene expression and morphology provides new insight into how microglia change with age | bioRxiv | from Prof. Stephen Quake's lab Stephen Quake Stanford University biorxiv.org/content/10.110… #MERFISH #microglia
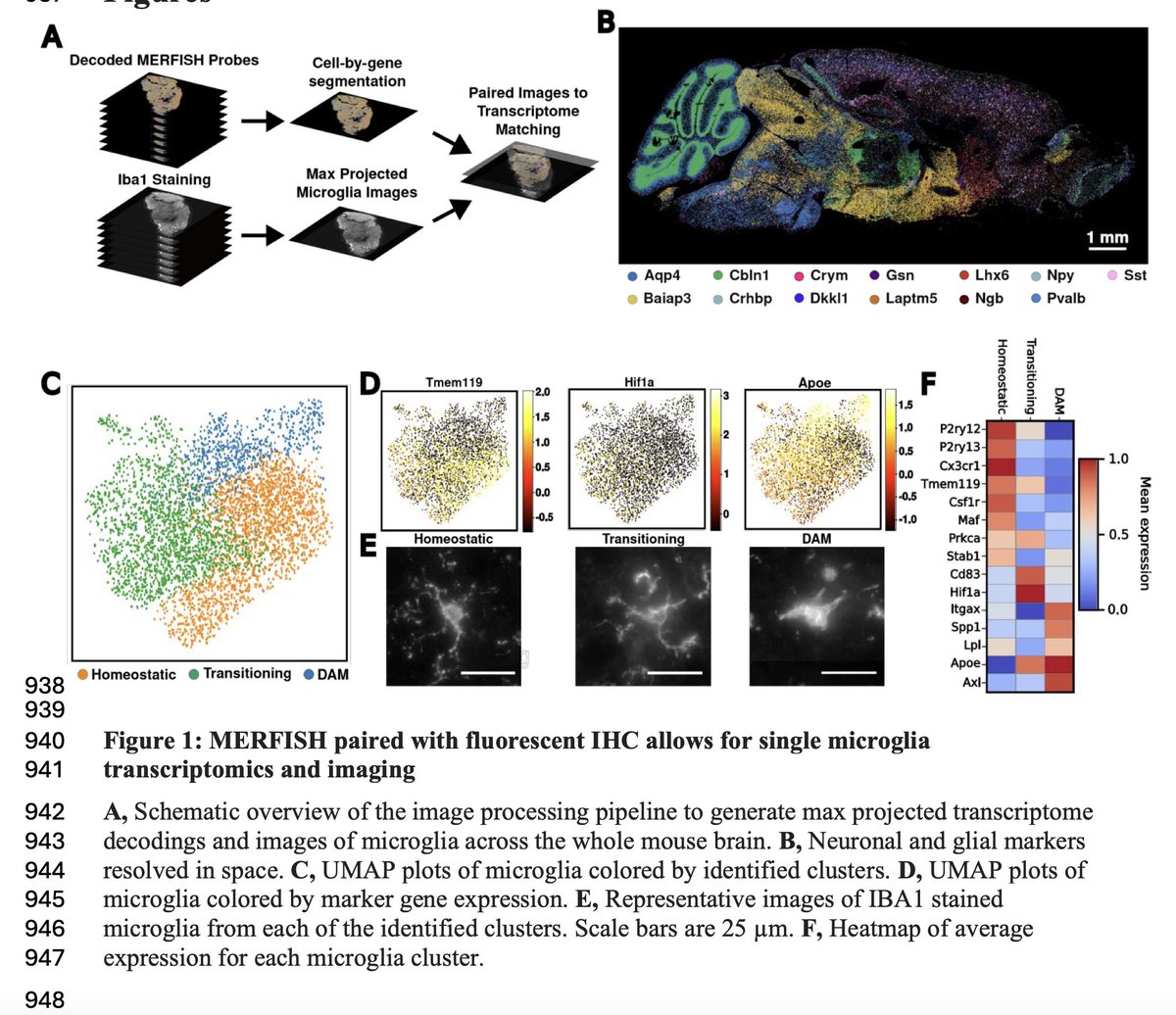





Systematic evaluation of single-cell multimodal data integration for comprehensive human reference atlas | bioRxiv | from Elisabetta Mereu 🇺🇦 and Joshua Levin labs biorxiv.org/content/10.110… #singlecell #scATACseq
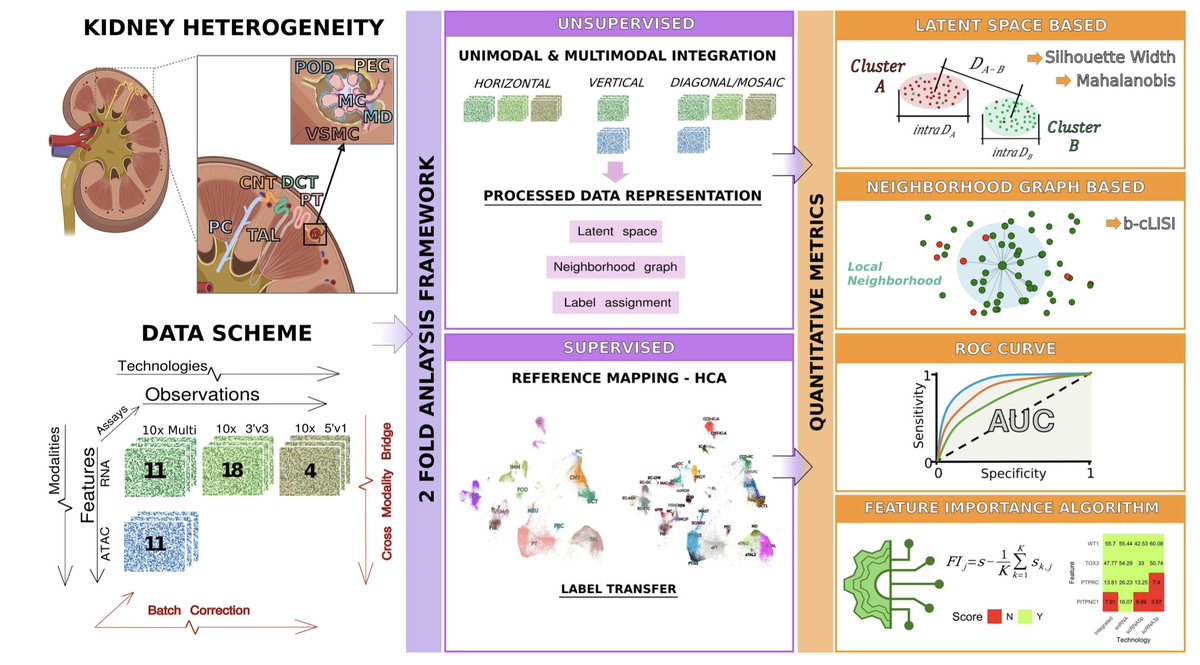

Scalable spatial transcriptomics through computational array reconstruction | Nature Biotechnology | from Prof. Fei Chen's Lab and their collaborators at Broad Institute Fei Chen Lab Chenlei Hu Broad Institute nature.com/articles/s4158…
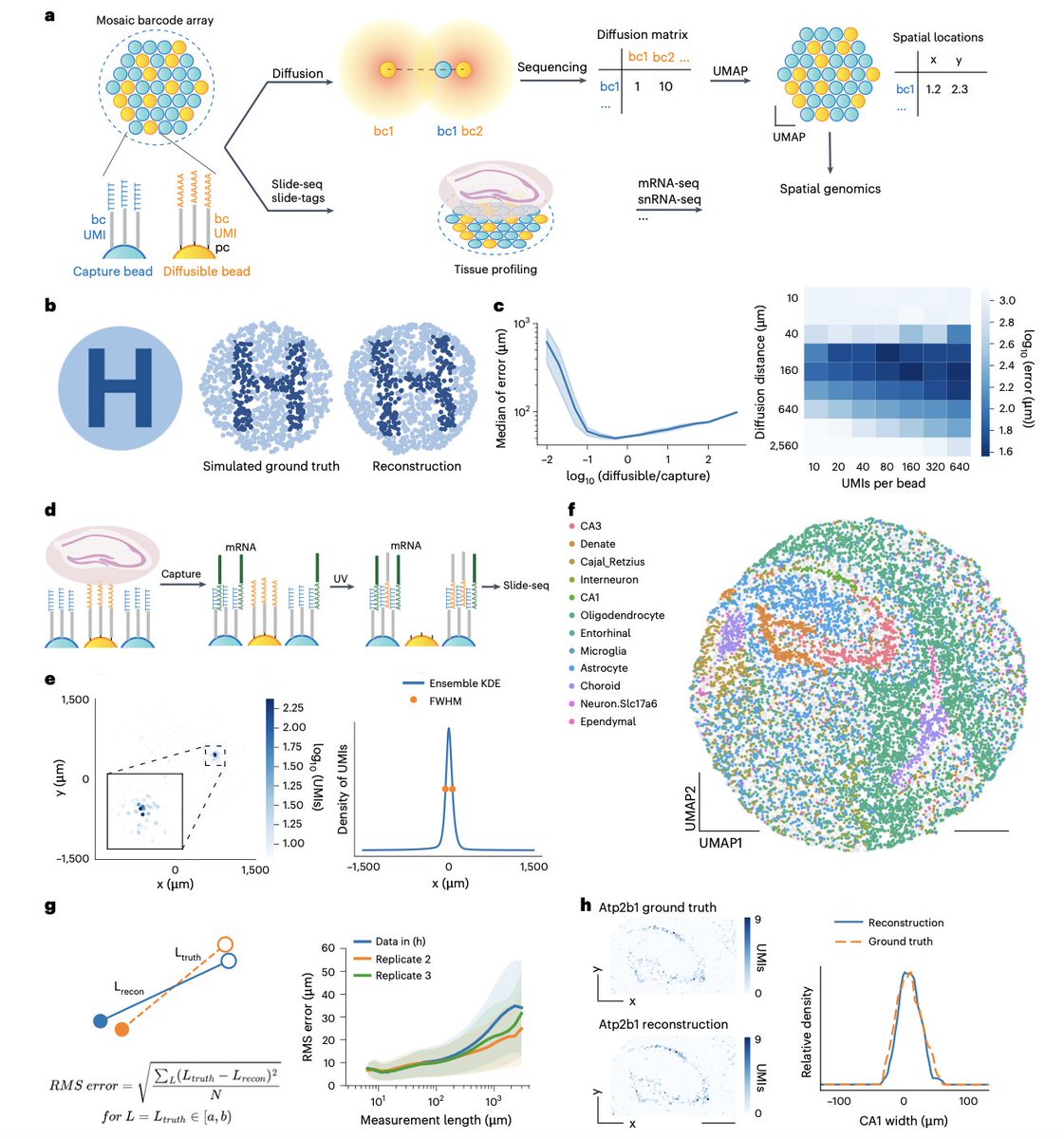


Predicting gene expression from DNA sequence using deep learning models | Nature Reviews Genetics Nature Reviews Genetics nature.com/articles/s4157…



📣 The recording of "Spatial Transcriptomics Technologies: A Primer" by Garam Kim is now available: youtube.com/watch?v=JbKCH7… This talk is part of Broad Institute's MPG Primer series. For more info, check out broad.io/MPGPrimer



AlphaGenome: advancing regulatory variant effect prediction with a unified DNA sequence model | from DeepMind Google DeepMind storage.googleapis.com/deepmind-media…










