Becca J. Carlson
@beccajcarlson
Associate @FlagshipPioneer • PhD @mit_hst @broadinstitute
• Immunology, virology, microscopy, functional genomics
ID: 1682144844192571392
http://linkedin.com/in/beccajcarlson 20-07-2023 21:46:03
128 Tweet
323 Followers
1,1K Following




Our paper on continuous mutagenesis in living cells using HACE is now out in Science Magazine! We now demonstrate that HACE continuously generates mutations over a 10-day period and enables targeting using dCas9 without introducing DNA nicks. science.org/doi/10.1126/sc…


In collaboration with Reuben Saunders, Jonathan Weissman's Lab, and Xiaowei Zhuang, we are very excited to release Perturb-Multi: a platform for pooled multimodal genetic screens in intact mammalian tissue. Check it out! biorxiv.org/content/10.110…

What if you could design binders for specific peptide-MHC complexes instead of hoping for hits from a screen? Bingxu Liu and team show we can! 🤩

Excited to see the work from Matteo Gentili Becca J. Carlson and others from our collab with the Hacohen lab Broad Institute out now!

1/ Thrilled to share an advancement in gene therapy from my PhD in Nature Communications! We've developed a new approach to reduce immune responses while maintaining efficiency—paving the way for safer, more effective therapies. Big thanks to Feng Zhang & Mirco J. Friedrich! bit.ly/redicas9


The unique pharmacology of targeted degraders (eg PROTACs) leverages their sub-stoichiometric “catalytic” activity. Now Lycia Therapeutics reports catalytic (cata)LYTACs for potent, durable extracellular TPD, cleared IgE in NHPs, superior to omalizumab 👏 biorxiv.org/content/10.110…

Excited to share the 1st of several stories dissecting TEs + chromatin in normal/malignant hematopoiesis. We identified histone demethylases KDM4A/C as vulnerabilities in DLBCL + discovered novel KDM4 inhibitors. Preprint👇 Broad Institute Boston Children's biorxiv.org/content/10.110…

Out now in Cell Reports our collab w/ Biederer Lab on image-based pooled genetic screening to identify regulators of synapse formation cell.com/cell-reports/f…


🎉Preprint alert! Our Ph.D. students Chujun He and Jiaqi Zhang , supervised by Eric and Wendy Schmidt Center Director Caroline Uhler and collaborator Munther Dahleh, have developed #MORPH, a MOdular framework for predicting Responses to Perturbational cHanges. Learn more ⬇️


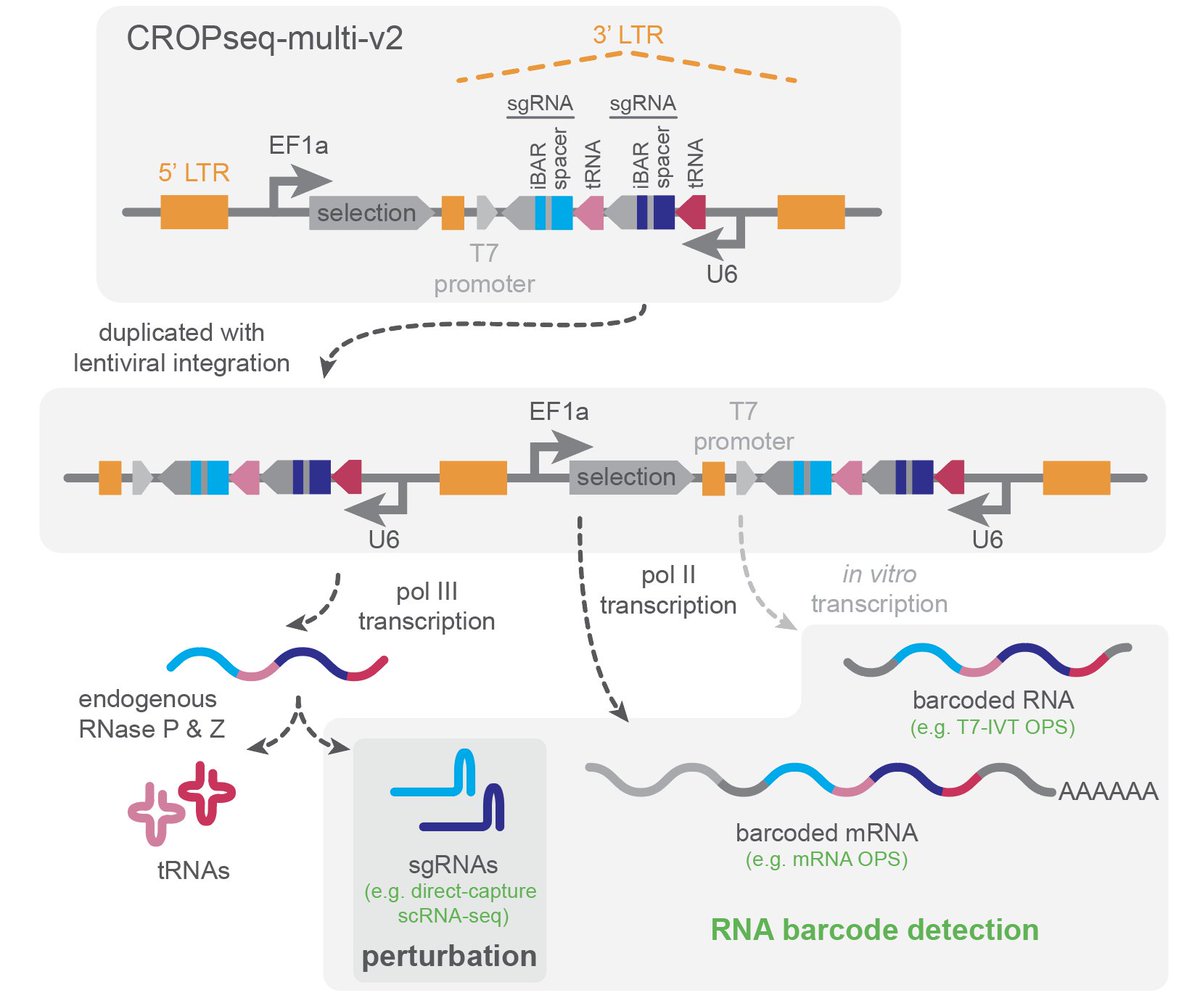
We’re Blainey Lab sharing a major update to CROPseq-multi, our versatile system for CRISPR screens that is compatible with individual and combinatorial perturbations, diverse SpCas9-based technologies, and multiple high-content, single-cell readouts. biorxiv.org/content/10.110…

ID possible new drug targets for treating Ebola infection via CRISPR screen, including UQCRB as a post-entry regulator of viral replication & STRAP as host regulator of equilibrium between viral RNA-protein Becca J. Carlson Blainey Lab nature.com/articles/s4156…


Our study on image-based genetic screens to identify novel host regulators of Ebola virus is now online Nature Microbiology! Thanks also to Broad Institute for writing a news story on our work: broad.io/Ebola-News-0724