
Claudio Alfieri
@calfieri84
ID: 975059782628532224
17-03-2018 17:22:22
482 Tweet
404 Followers
516 Following

SPACEtomo is officially published! :P nature.com/articles/s4159… I'm so grateful for all the support I received especially from Rado Danev and everyone who shared their data with me! And of course thanks to David Mastronarde and the amazing #SerialEM for making it possible!





🚨Preprint, RT appreciated🙏 Our work ‘Substrate recognition by human separase’ by jun, Sophia Schmidt, Margherita Botto UNIGE_en Université de Genève Molecular & Cellular Biology Dept 🔬 Geneva 🇨🇭 Dubochet Center for Imaging (DCI) and members from the Morgan UC San Francisco and Francis O'Reilly labs is online. 🔗biorxiv.org/content/10.110…

Passionate about structural biology and #biochemistry? Join David Haselbach’s group through the Vienna BioCenter #PhD Program (Vienna BioCenter Scientific Training)! ⏳ Deadline: April 15, apply now: training.vbc.ac.at/phd-program/ 📷Watch David's 'Scientist Snapshot': youtube.com/watch?v=DXTOW3…



#NewPaper 🧬 A new review dives into the structural biology of tRNA modifying enzymes. Authored by our scientists led by Sebastian Glatt highlighting how cryoEM & crystallography reveal mechanisms behind key modifications. Read here: shorturl.at/akpeB Uniwersytet Jagielloński



Our lab collaborated with BYU_BiophysicsGroup to create this important tomogram dataset, which will be a useful benchmark for future machine learning developments in the cryo-ET world.

📣 New in The EMBO Journal : Our researchers lead by Sebastian Glatt , together with @IIMCB_Warsaw, Durham University & Synchrotron SOLEIL reveal how #pseudouridine boosts human #tRNA stability using cryo-EM & modeling. Big leap for RNA-based therapies! Uniwersytet Jagielloński SOLARIS Centre





Check out our latest study NatureStructMolBiol: Establishing a consensus model for #ubiquitin chain assembly by HECT #E3 ligases:#cryoEM structures of #TRIP12 forming K29-linked and K29/K48-branched chains! ❕Publication: nature.com/articles/s4159… Samuel Maiwald Universiteit Leiden

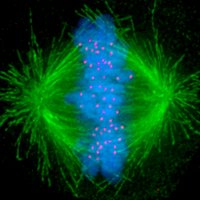

A transcription balance required for genome stability 🧬⚖️ Our work on RNA Pol II transcription speed control by the DNA helicase RECQL5 and the transcription-coupled DNA repair complex is now on NatureStructMolBiol #CryoEM #transcription nature.com/articles/s4159…




