
Chris Bailey
@cbailey_58
Budding haematologist and PhD student at the Crick looking into Cancer Evolution. Funded by CRUK.
ID: 1069727353234960385
03-12-2018 22:57:30
150 Tweet
418 Followers
516 Following

🔥Excited to share Mădălina Giurgiu Decoil out today Genome Research If you have Nanopore genome sequencing and want to study the structural heterogeneity of ecDNA, try it. Thank you to all the collaborators Cancer Grand Challenges team eDyNAmiC, Max Delbrück Center Charité - Universitätsmedizin Berlin Memorial Sloan Kettering Cancer Center

🚨 Excited to share our latest paper in Cell identifying how under-replicated chromosomes in micronuclei become shattered by the Fanconi anemia pathway during mitosis. This work was led by UTSW Graduate School of Biomedical Sciences student Justin Engel. 🧵 (1/13): cell.com/cell/fulltext/…

Together with Charles Swanton, delighted to share that our new tool, MHC Hammer - which can detect different types of HLA disruption - mutation, loss, RNA repression and alt. splicing - is now out in #NatureGenetics and available to use…nature.com/articles/s4158…... 🧵below



⚪️🧬Un macroestudio liderado por Oriol Pich (#IRBalumni, ahora en The Francis Crick Institute) y Chris Bailey, revela diminutos círculos de ADN en las células de pacientes con algunos tipos de #cáncer, como el de mama #HER2+ —uno de los más agresivos— y el #glioblastoma en el cerebro. 🗣️






🚨So proud to present the first study from our Computational Cancer Genomics (CCG) Research Group group together with Charles Swanton Nnennaya Kanu and led by Olivia Lucas with key collaboration Sophie Ward and fundamental support Rija Zaidi Abi Bunkum & many others Out in Nature Genetics 👇 doi.org/10.1038/s41588… 🧵👇




🚀 Our paper introducing ImmuneLENS, a new tool that measures T and B cell fractions from WGS data. This builds on our previous method, T cell ExTRECT, that used a signal from V(D)J recombination to measure T cells. Out today in Nature Genetics doi.org/10.1038/s41588… 🧵👇



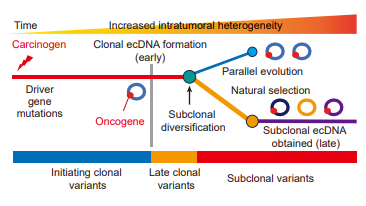
Now online in Cancer Discovery: Spatial-Temporal Diversity of #ecDNA Shapes Urothelial Carcinoma Evolution & Tumor-Immune Microenvironment - by Wei Lv, Yuchen Zeng, Conghui Li, Yuan Liang, Birgitte Regenberg, Yonglun Luo, Chunhua Lin, Peng Han, and colleagues doi.org/10.1158/2159-8…



A beautiful study led by my brilliant colleagues Oriol Pich Elsa Bernard that opens up the impact of tumour infiltrating clonal haematopoiesis
