Francesco Beghini
@chocophlan
Postdoctoral Fellow at @Yale | Human microbiome | Bioinformatics | Go @Patriots!
ID: 93675118
30-11-2009 17:34:25
5,5K Tweet
367 Followers
752 Following


I wrote a blog post about scientific racism and the AllofUsResearch figure illustrating their ancestry analysis. The post was inspired by a presentation of Delaney Sullivan at our lab DEI meeting organized by Nikhila Swarna (Nikki) last week. liorpachter.wordpress.com/2024/02/26/all… cc Josh Denny



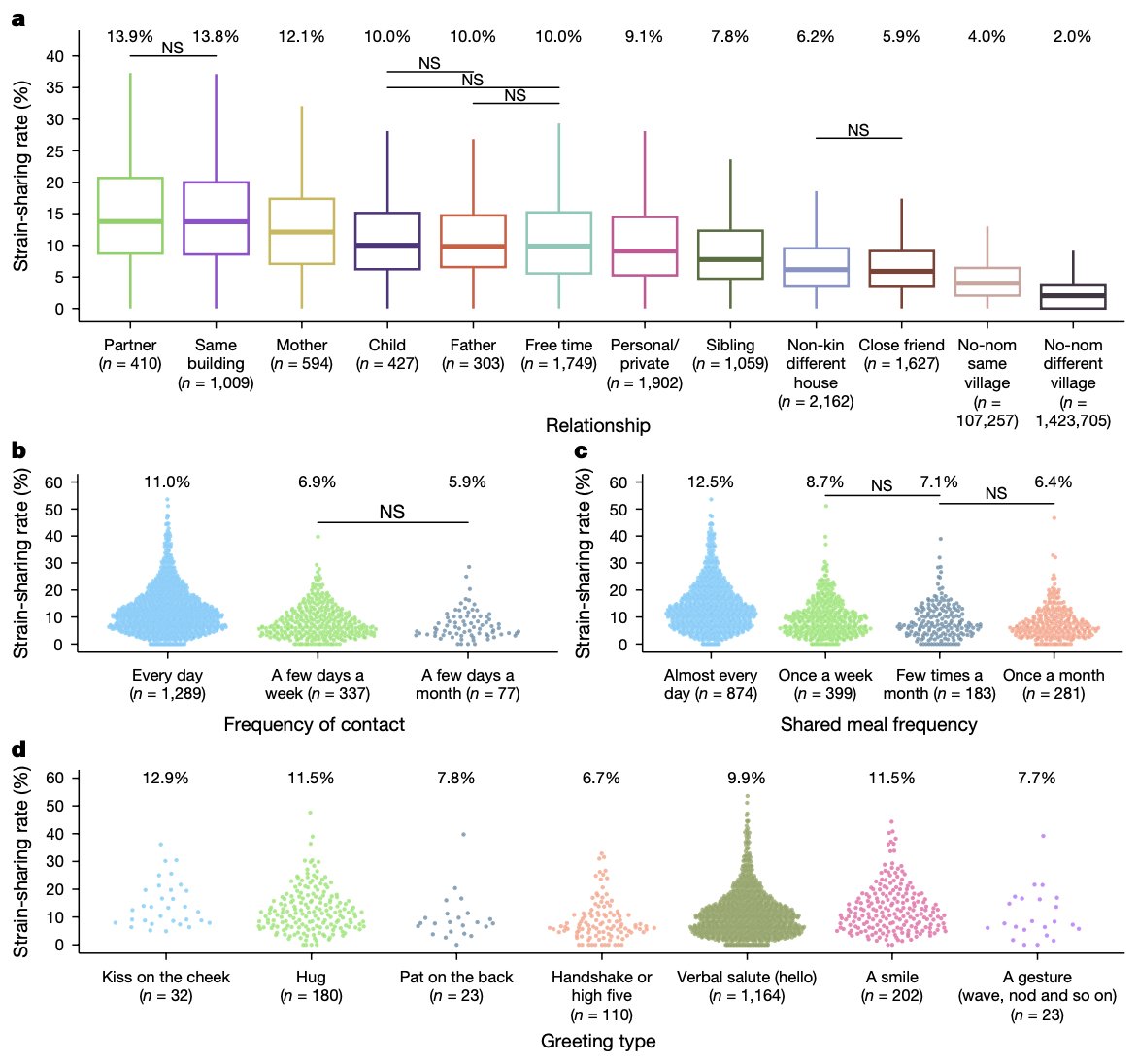
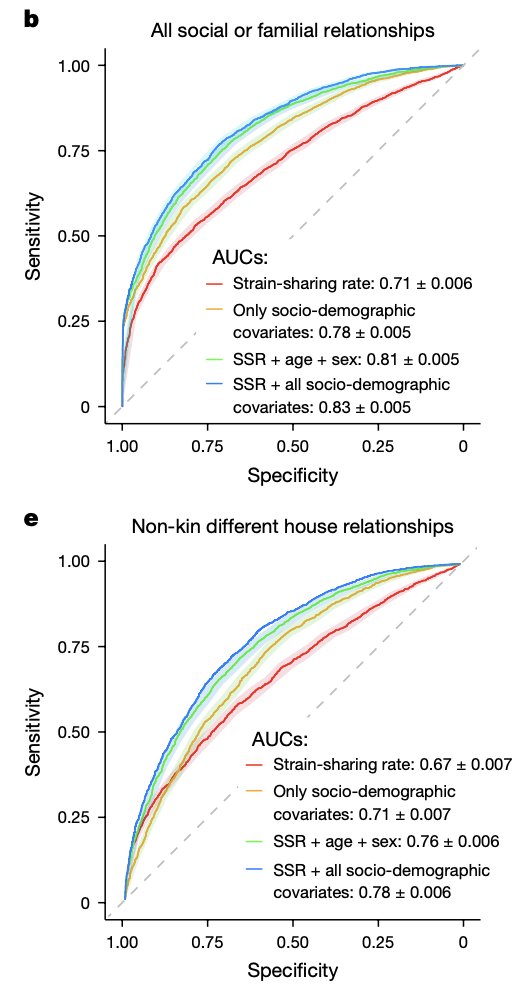
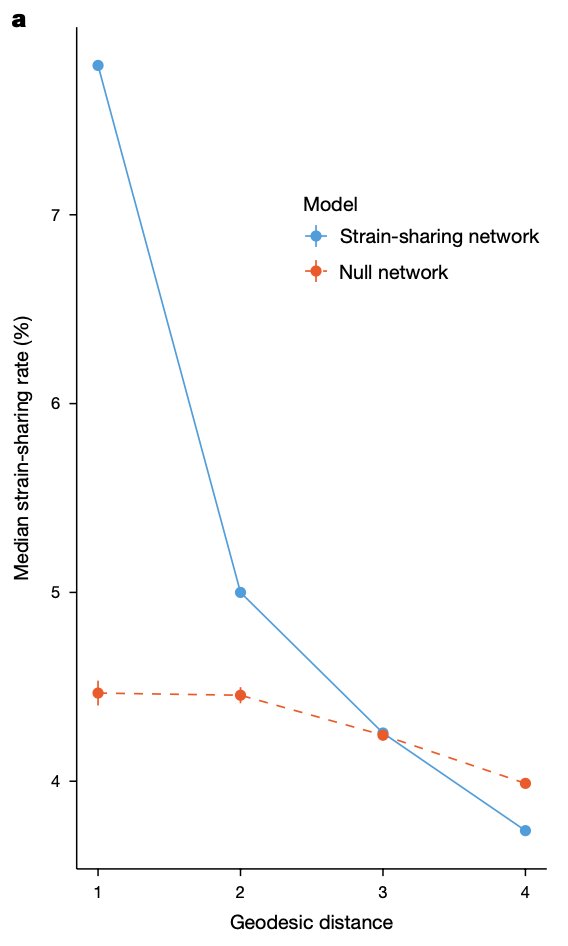
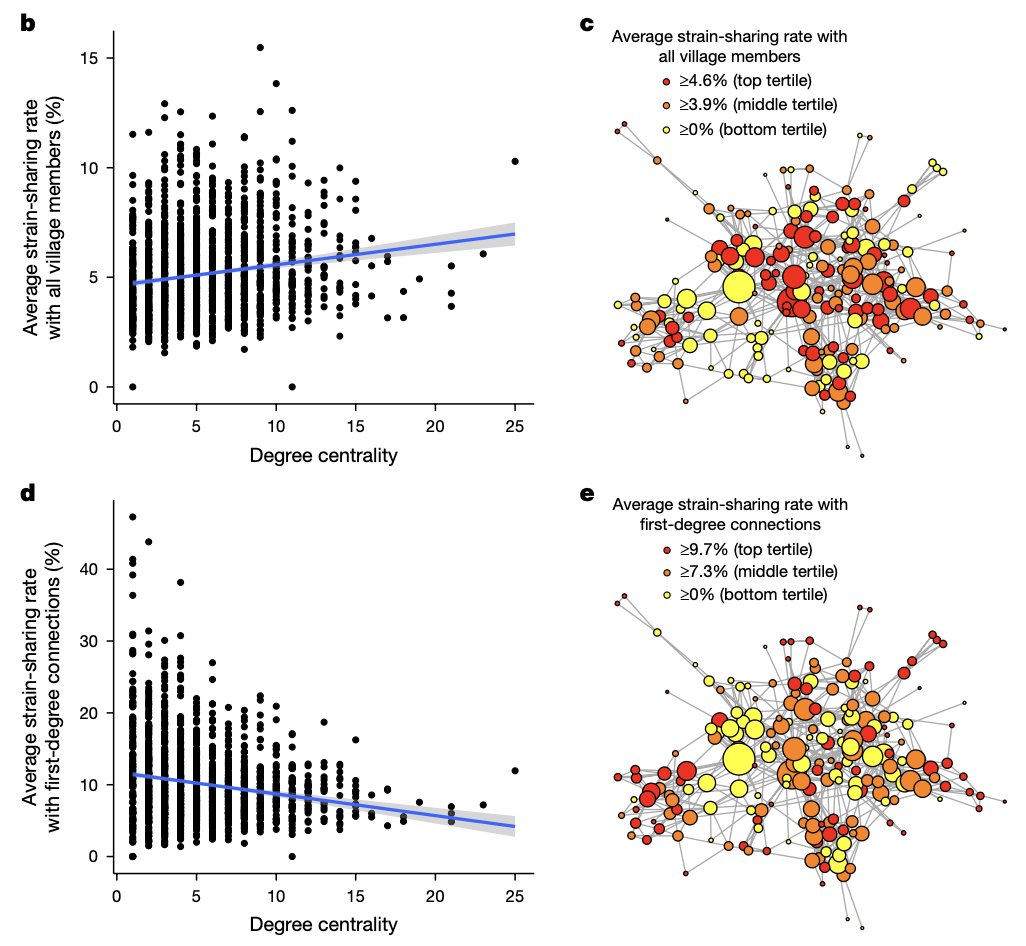
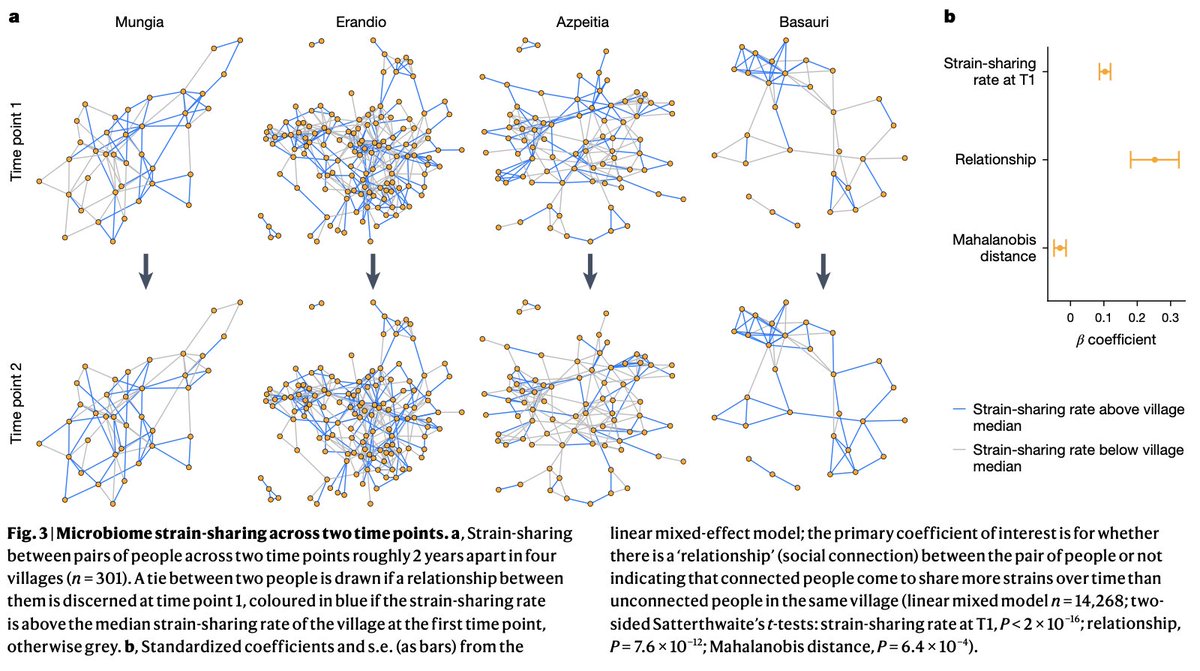
Environmental, socioeconomic, and health factors are associated with gut microbiome species and strains in isolated Honduras villages. New #HNL work with Shiv, Francesco Beghini Marcus Alexander, MD, PhD A Singh, R Juarez, & Ilana Brito cell.com/cell-reports/p… #microbiome #LMICs




"Gut microbiome strain-sharing within isolated village social networks" nature.com/articles/s4158… with Francesco Beghini, Jackson Pullman, Marcus Alexander, MD, PhD, Shiv, Drew Prinster (same @ other app), Adarsh Singh, RM Juárez, Edo Airoldi, Ilana Brito #HNL 2/











This work was supported by the NOMIS foundation, with additional support via Schmidt Futures Eric Schmidt Pershing Square Philanthropies Bill Ackman JM Rothberg. Underlying cohort supported by Gates Foundation & NIH. New support for work on the biology of social life at #HNL via Paul Graham. 19/

A new study found that we share parts of our microbiome with people in our social networks, beyond family members. Dr. Nicholas A. Christakis joins us to discuss the research and how scientists can identify your friends—just by looking at your poop. buff.ly/3VphYOQ