
Cody Heiser
@cody_heiser
Computational systems biologist working in pharma/biotech. Longhorn alum.
ID: 452679071
https://codyheiser.github.io/ 02-01-2012 03:58:10
805 Tweet
157 Followers
175 Following




#WinterClassic is over but hockey at Cotton Bowl is not! The UT (Texas Ice Hockey) vs. A&M (Texas A&M Ice Hockey) rivalry will be renewed, except it'll be on ice! A big deal for TCHC Hockey/ACHA Men's D2 and hockey in TX. My latest all about it The Athletic Dallas👇🏽 theathletic.com/1475250/2020/0…

Throwback to last week’s QSBC seminar with Cody Heiser presenting - our first of the semester! Looking forward to another semester of seminars, VU speakers, and visiting speakers
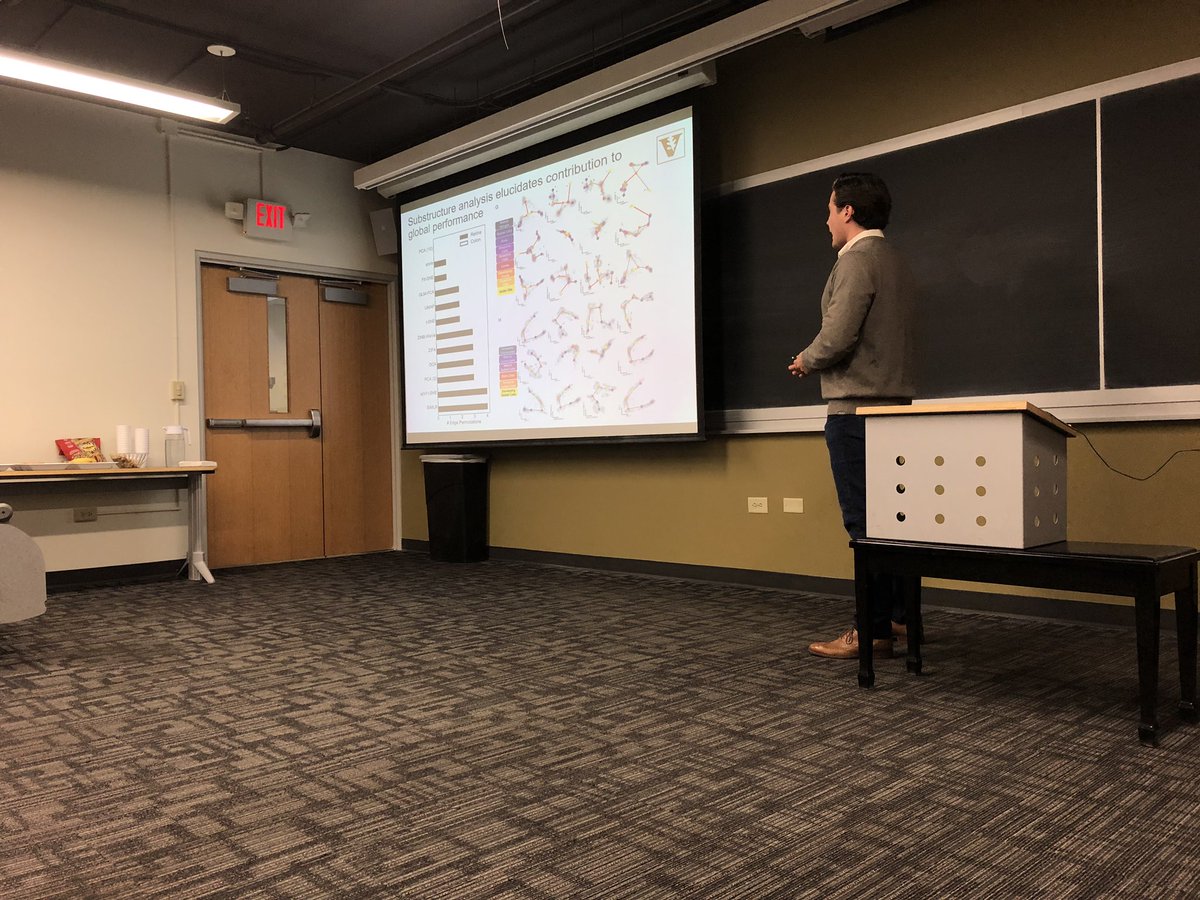

We routinely use various dimension reduction tools to project multi-dimensional datasets into latent spaces for visualization and downstream analysis, but how much information do we lose? Cody Heiser may provide some insight. cell.com/cell-reports/f… [1/4]

We present dropkick - a machine learning Python package that leverages ambient signal for QC and automatic droplet filtering of scRNA-seq data Cody Heiser biorxiv.org/content/10.110…


Our dropkick cell filtering manuscript is now online at Genome Research. dropkick provides fast, automated cell identification in droplet-based #singlecell RNA-seq datasets, performing especially well on high-background samples. dx.doi.org/10.1101/gr.271…




Cell Vanderbilt CPB Cherie Scurrah Vanderbilt Cell and Developmental Biology A cytotoxic immune microenvironment in hypermutated tumors confer better patient prognosis and immunotherapy response. Strikingly, serrated polyps (precursors to MSI-H CRCs) already present a cytotoxic immune microenvironment preceding hypermutation



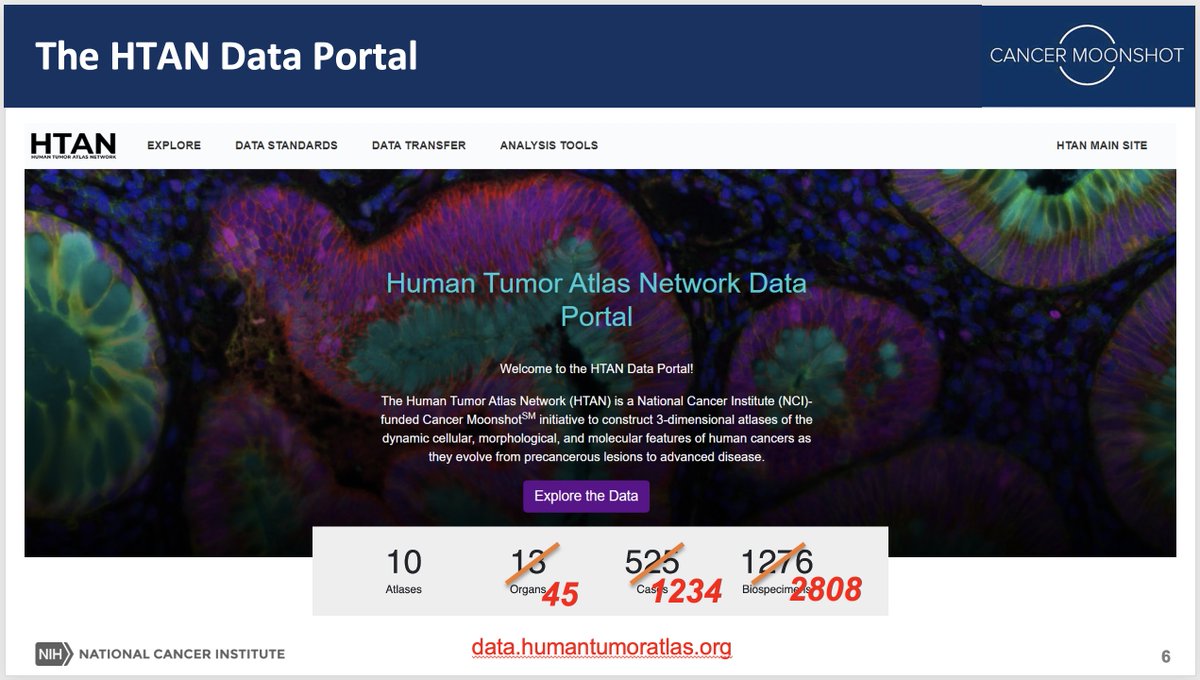
Outstanding kickoff for the annual HTAN meeting! Thanks to Ken Lau Lab William J. Greenleaf @SantagataLab Michael Snyder, PhD Reyka Jayasinghe, PhD Nataly Naser Al Deen Cody Heiser Great to see a talented and diverse group of scientists generating incredible approaches to spatially resolved data! @NCIHTAN

First preprint by LauLab student Harsimran Kaur about MILWRM - a python package that identifies consensus tissue domains across specimens (aka slides) in high dimensional mIF and ST data. Looking forward to your QE! Simon Vandekar Cody Heiser @etmckinley biorxiv.org/content/10.110…


Capstone paper from my dissertation research is out in Cell! Thanks Ken Lau Lab and Vanderbilt CPB for the tremendous support along the way.



Unsupervised pixel-level spatial analysis tool from my colleagues at Vanderbilt! Simple, fast, interpretable - very promising for all flavors of ST and MxIF data. Great work by Harsimran Kaur


