Cyrus Tau
@cyrus_tau
phd student and nsf fellow @ harvard bbs | prev: bioe @ ucb, arc institute, hsu lab
ID: 1349262450294620160
13-01-2021 07:50:26
131 Tweet
148 Followers
186 Following




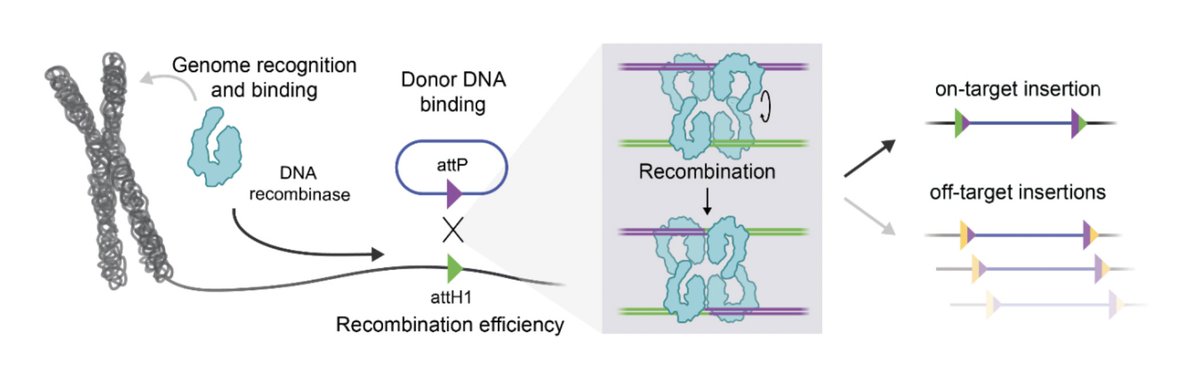
Excited to share this new work from Arc Institute-- led by graduate student Sita Chandrasekaran and undergrad Cyrus Tau with Patrick Hsu. Building on our previous work on Cas13d-based RNA targeting we can now write full sequences into endogenous RNA!


Long non-coding RNAs comprise a large part of the genome, yet loss of one copy (haploinsufficiency) of a lncRNA is not known to cause a human disease. Read our preprint to learn how deletions of the lncRNA CHASERR cause a severe neurodevelopmental disorder.medrxiv.org/content/10.110…

Nice story in Endpoints News on our lab's RNA writing work by Ryan Cross the Science Boss, alongside complementary studies from friends and colleagues!



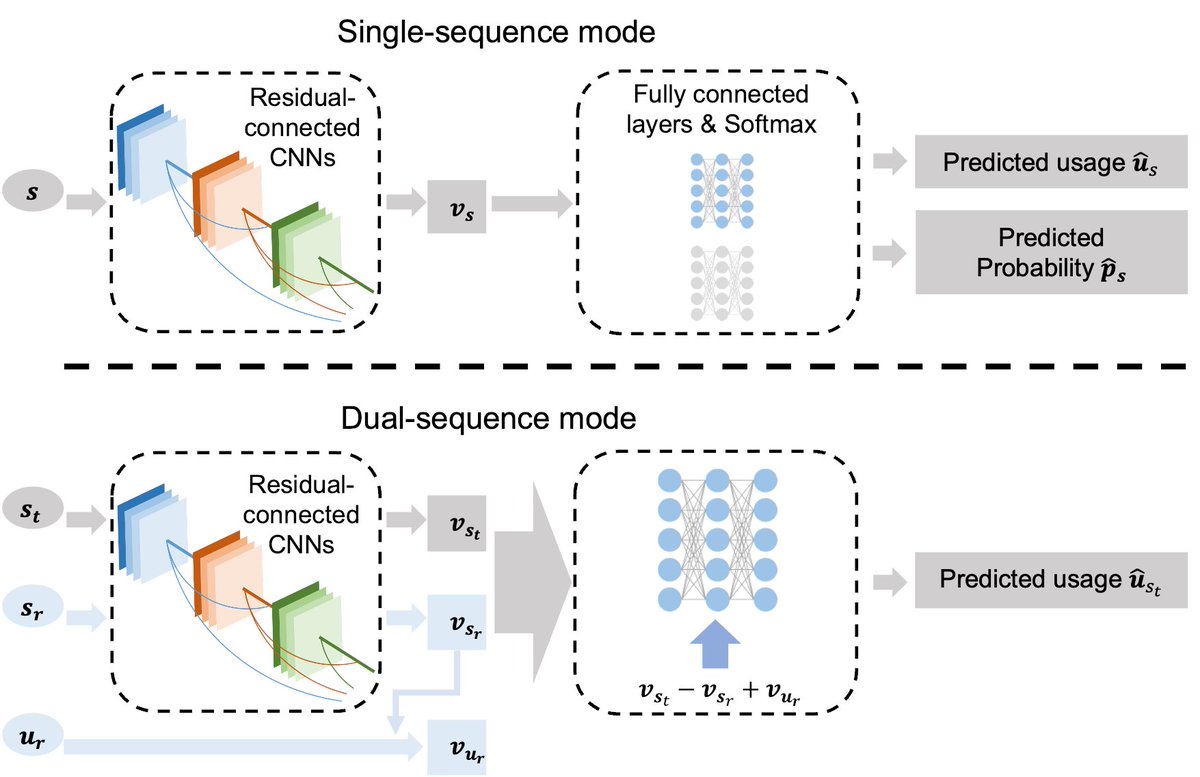
Our most recent preprint on #DeltaSplice for quantitative prediction of splicing and splicing-altering mutations. Great collaboration with Tao Jiang Lab. Led by Chencheng Xu@Jiang lab and a former postdoc Suying Bao in my lab. Software available soon-stay tuned!

Excited to share our work published in Science Magazine today answering the question: “Where does Cas13 come from?” science.org/doi/10.1126/sc…