David Koes
@david_koes
Removing barriers to computational drug discovery one bit at a time.
Patiently waiting for scientific twitter to migrate somewhere (anywhere) else.
ID: 3427962639
http://bits.csb.pitt.edu 17-08-2015 12:44:26
778 Tweet
2,2K Followers
438 Following





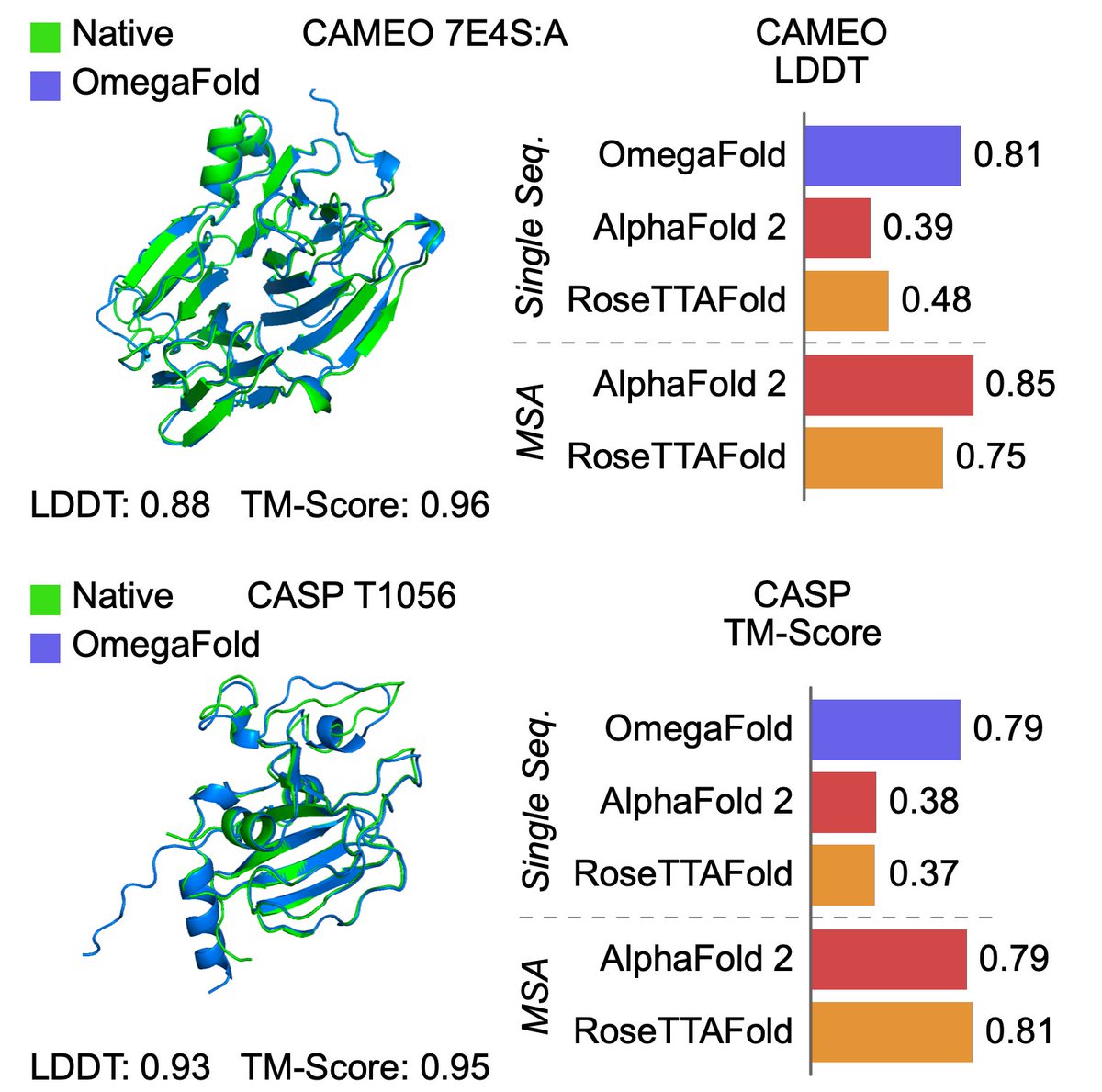
Protein structure can be predicted from a single sequence alone with high accuracy. Helixon Bio team have developed OmegaFold, achieving performance similar to RF and AF2's MSA versions. Only a single sequence is given as input. 1/5


Thanks to Andrew McNutt (David Koes group), who converted #GNINA’s models’ weights from #Caffe to #PyTorch, 🔥 gnina-torch 🔥 (a PyTorch implementation of GNINA’s scoring function) now ships with all pre-trained GNINA’s models! 🧵 1/6 github.com/RMeli/gnina-to…



🔥 gnina-torch 🔥 is now available Hugging Face. Just drag-and-drop your protein-ligand complex and score it with #GNINA pre-trained models! Try it out: huggingface.co/spaces/RMeli/g… Powered by Hugging Face Gradio PyTorch pytorch-ignite 3Dmol.js (David Koes) 🧵 1/4


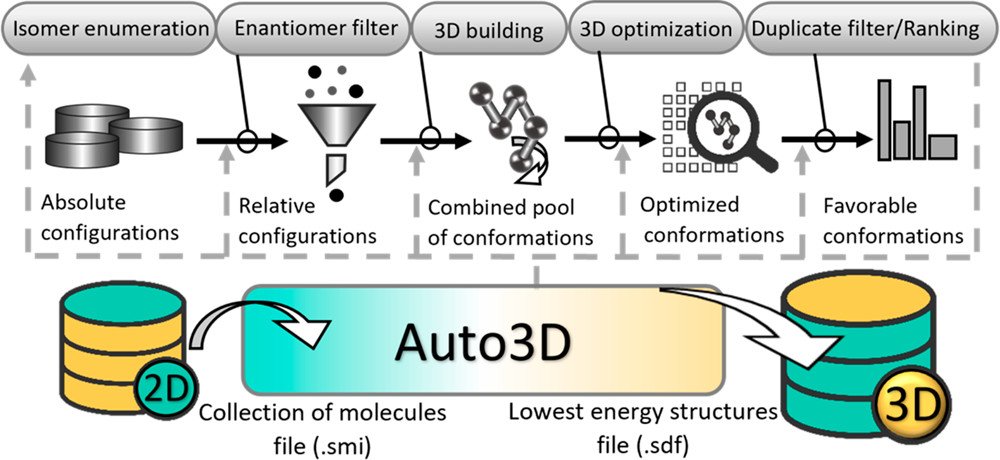
A new paper just published in JCIM & JCTC Journals! Auto3D: Automatic Generation of the Low-Energy 3D Structures with ANI Neural Network Potentials. Collaboration with Adrian Roitberg 🏳️⚧️ #compchem #MachineLearning pubs.acs.org/doi/full/10.10…


Excited to share BigBind: Learning from Nonstructural Data for Structure-Based Virtual Screening doi.org/10.26434/chemr…. With Paul Francoeur, Rishal Aggarwal, Konstantin Popov, David Koes, and Alex Tropsha


RT @GabriellaGerla1: TECBio REU @ Pitt Applications are open now!! This is a great program to do computational biology research during undergrad, and determine if you want to pursue a PhD. tecbioreu.pitt.edu pic.x.com/2qNwL26Vyw




Excited to share PLANTAIN: arxiv.org/abs/2307.12090. Our docking approach applies diffusion-inspired training to the classic idea of pose score minimization. We achieve state-of-the-art results while being 10x faster than others. W/ Konstantin Popov, David Koes, and Alex Tropsha