Davide Bolognini
@davidebolo93
Senior Bioinformatician @humantechnopole, Population & Medical Genomics Unit.
ID: 4885447870
https://github.com/davidebolo1993 07-02-2016 18:07:26
156 Tweet
107 Followers
69 Following

Very happy to share the first of many stories we've been working on here at Human Technopole



Congrats Gaia Pigino and team 🔥




Long read Oxford Nanopore data from 1k genome project - analysed and also available for everyone to have fun with! search.app/cLvwtcVYwuVFpE…



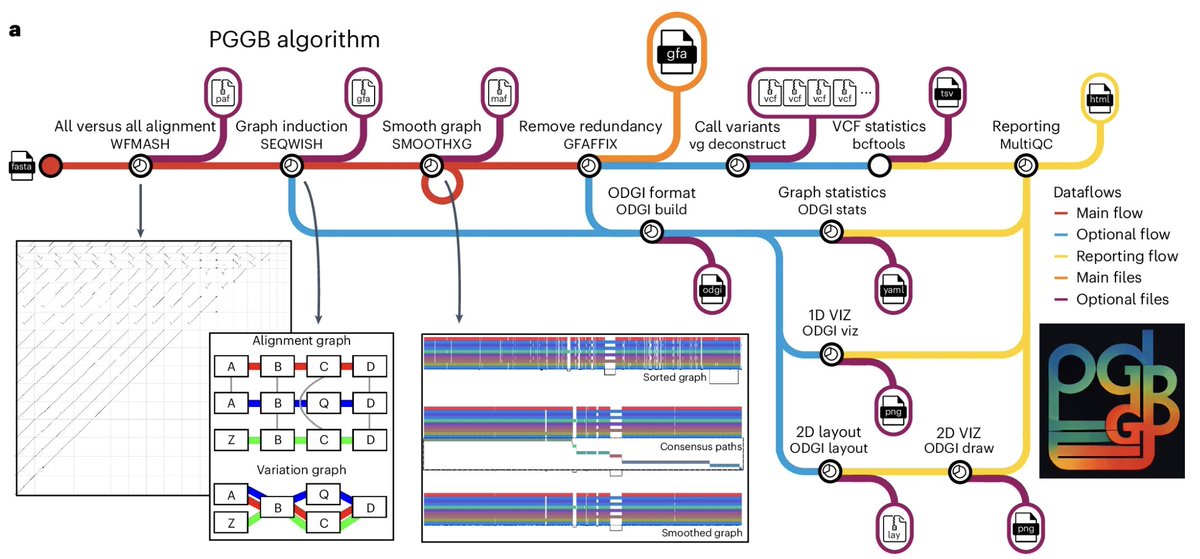
PanGenome Graph Builder is a modular framework for efficiently building unbiased pangenome graphs, supporting diverse downstream analyses. Erik Garrison Andrea Guarracino nature.com/articles/s4159…

At #ASHG and interested in deep learning for disease prediction? Come visit Francesco Cisternino ‘s poster: imaging + transcriptomics to understand how carotid plaque morphology affects patients symptoms and MACE risk (board 4081T) #ASHG2024


Thank you Sara! Happy to finally present the work we’ve been doing with the labs of Sander van deer Laan (UMC Utrecht) and Clint Miller (UVA) on carotid plaque morphology and vulnerability at #ASHG24 😊



🎙️ Per Tgr Leonardo, Nicole Soranzo racconta il progetto Genome of Europe, il più grande progetto genomico finanziato dall'UE, per promuovere prevenzione e cure personalizzate. (da min 11.20) rainews.it/tgr/rubriche/l…

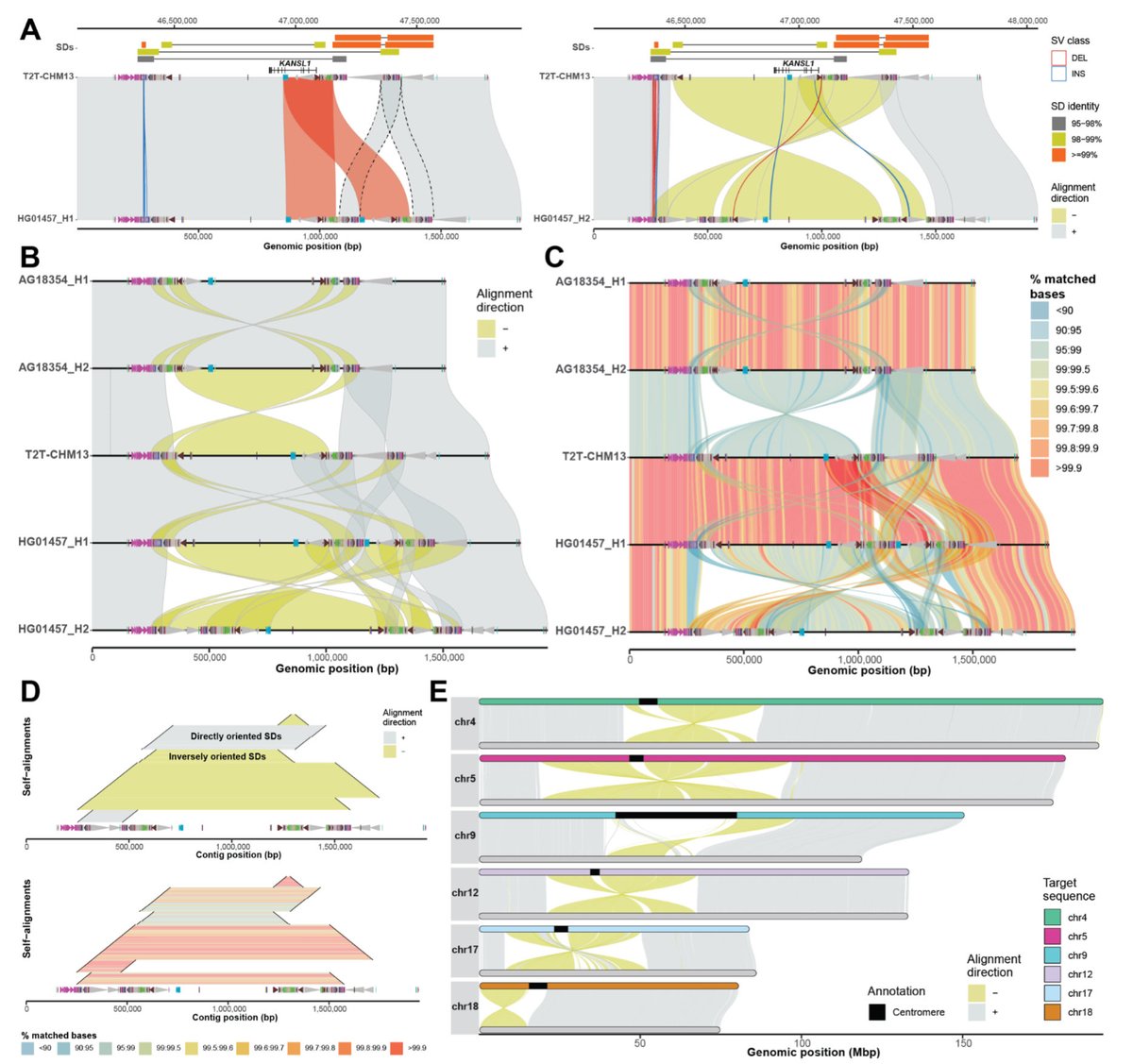
Our Cell Genomics preview “Double or Nothing – Ancient Duplications in the Amylase Locus Drove Human Adaptation” covering Yilmaz F. (Feyza Yilmaz) et al., Science 2024 and Bolognini D. (Davide Bolognini) et al., Nature 2024 is now out. massilani-lab.com/new-publicatio…



So excited that our team was awarded an #ERCAdG grant European Research Council (ERC) this year. A testament to the brilliance, hard work and dedication of my present and past teams Human Technopole Wellcome Sanger Institute Cambridge University. 🤩🤩