D.Sherpa
@dawafutisherpa
Biochemistry-structural biology
ID: 3246001367
11-05-2015 07:56:48
93 Tweet
185 Followers
358 Following

How does multisite #phosphorylation of an #E2 enzyme dictate its pairing with a multisubunit #E3 #ubiquitin ligase to regulate metabolism? Check out our story available now in Molecular Cell! ➡️ cell.com/molecular-cell… D.Sherpa Schulman lab Max Planck Institute of Biochemistry (MPIB)



How does multisite #phosphorylation configure the catalytic E2-E3 assembly for metabolic enzyme targeting? Check out our story available now in its final version in Molecular Cell! ❕cell.com/molecular-cell… Jakub Chrustowicz D.Sherpa #ubiquitin

Excited to share our new preprint. Using #cryoEM 🔬we obtained multiple snapshots 📸 of the chaperone-mediated multistep assembly pathway of the human 20S #proteasome. Max Planck Institute of Biochemistry (MPIB) @HarvardCellBio biorxiv.org/content/10.110…

See how CRLs forge ubiquitin chains on substrates - and degrader-recruited neo-substrate - at high-res! Millisecond polyubiquitylation! #cryoEM #TPD NatureStructMolBiol Max Planck Institute of Biochemistry (MPIB) UNLV College of Sciences 👉shorturl.at/eFS68 Related exciting work from Alessio Ciulli shorturl.at/hNSU5


🚨Excited to share that our manuscript is now published in NatureStructMolBiol. Using #cryoEM 🔬we obtained multiple snapshots 📷 of the #chaperone-mediated multistep assembly pathway of the human 20S #proteasome. Max Planck Institute of Biochemistry (MPIB) @HarvardCellBio Aligning Science Across Parkinson’s nature.com/articles/s4159…

Excited by #ubiquitin & #proteomics? Openings for joint doctoral/postdoc positions at Matthias Mann Lab and the Schulman groups Max Planck Institute of Biochemistry (MPIB). Employ cutting edge mass spectrometry to decode E3 ligase networks & signaling pathways in critical biological areas. Apply now! #ScienceCareers

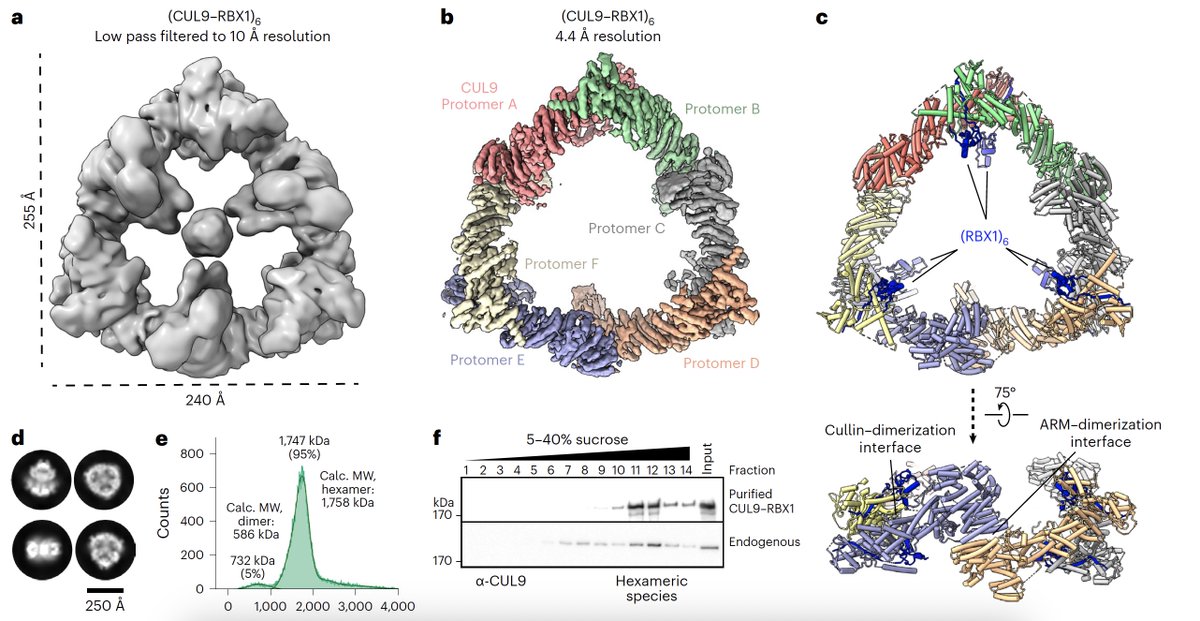
Wow! CUL9 is a GIANT E3 ubiquitin ligase! See unique cullin-RING-RBR all-in-one. +/- NEDD8. Now online in Nature Structural & Molecular Biology by Schulman Department ❕nature.com/articles/s4159… #ubiquitin #E3ligase #cryoEM @DHornGhetko @LinusHopf Matthias Mann Lab NatureStructMolBiol

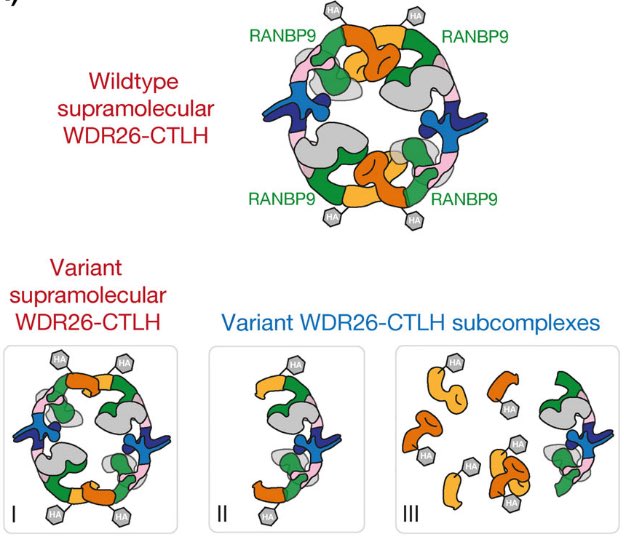
Check out our story revealing exciting regulation of the NAD+ biosynthetic enzyme NMNAT1 by the supramolecular CTLH E3 ligase assembly out in Molecular Cell! ➡️cell.com/molecular-cell… Karthik Gottemukkala, D.Sherpa, Arno Alpi Schulman lab Max Planck Institute of Biochemistry (MPIB)

Alpi, Schulman, Mann and Murray labs decipher regulation of human NMNAT1 (NAD+ biosynthetic enzyme) by gigantic WDR26-CTLH E3 ligase complex! ❕cell.com/molecular-cell… #ubiquitin #E3ligase Karthik Varma Gottemukkala Jakub Chrustowicz D.Sherpa Arno Alpi Matthias Mann Lab Peter J Murray


Congratulations to all our wonderfully talented young researchers honoured with an Otto-Hahn-Medal today for their outstanding scientific contributions - we're so proud of you🥳! Keep paving the way for a bright future!🤗Group photo of the winners with Patrick Cramer📸in Berlin☀️


Extremely grateful and honored to receive the Otto Hahn Medal from the Max Planck Society Max Planck Society . Thank you Brenda Schulman Max Planck Institute of Biochemistry (MPIB) , Arno Alpi , Jakub Chrustowicz , past and present GID team members and the Schulman Lab family for making this possible.



It official! The Hoyer lab has opened it's doors at the Van Andel Institute (VAI) in VAI Department of Neurodegenerative Science. For more information check out our website: hoyerlab.vai.org This is such an exciting adventure. I am thankful to my family, friends, and mentors who have helped me get here!

Congratulations to Anna Uzonyi of Weizmann Institute & Lukas Teoman Henneberg of Max Planck Institute of Biochemistry (MPIB), winners of the Amon Young Scientist Awards. They’ll present at the Amon Awards ceremony at the KI on 11/14 at 2 p.m. The MIT community & Amon Lab alumni are invited. mit-ki.org/4g5Gb5P


We are happy to share our new preprint bioRxiv "Unveiling the hidden interactome of CRBN molecular glues with chemoproteomics" led by Kheewoong Baek and Rebecca Metivier of Fischer Lab biorxiv.org/content/10.110…