
DDX3X Support UK
@ddx3xsupportuk
If you're a parent/carer please join our closed FB group, DDX3X Support UK, for support, chats and information with other UK and European parents/carers
ID: 847798191101800448
31-03-2017 13:10:14
49 Tweet
66 Followers
32 Following

1. Really excited to share our work using saturation genome editing to generate a variant effect map of the gene DDX3X. nature.com/articles/s4146…. It’s been a privilege to lead this work together with HongKee TAN, with brilliant mentorship from Matt Hurles and @GeretySebastian. A 🧵:

HongKee TAN Matt Hurles @GeretySebastian 2. Loss of function variants in DDX3X are the most common monogenic cause of neurodevelopmental disorders (NDD) in females. Somatic DDX3X mutations are known drivers of diverse cancer types, but whether DDX3X acted as a tumour suppressor or an oncogene was unclear.


HongKee TAN Matt Hurles @GeretySebastian Greg Findlay Lea Starita Jay Shendure 4. We used SGE in HAP1 cells to test the functional effect of >12,500 DDX3X genetic variants including all coding single nucleotide variants, all codon deletions, every coding small indel seen in clinical and population cohorts and all intronic SNVs near exons.

HongKee TAN Matt Hurles @GeretySebastian Greg Findlay Lea Starita Jay Shendure 5. DDX3X is essential for HAP1 cell growth. Therefore, to identify which genetic variants impair the function DDX3X we compared the relative abundance of the different genetic variants in culture over 3 weeks. Variants which impair the function of DDX3X become depleted over time.


HongKee TAN Matt Hurles @GeretySebastian Greg Findlay Lea Starita Jay Shendure 6. We identified 3,432 functionally abnormal variants in three different classes: fast-depleting, slow-depleting and enriched variants.


HongKee TAN Matt Hurles @GeretySebastian Greg Findlay Lea Starita Jay Shendure 8. Putative loss-of-function variants (fast and slow depleting variants) tended to be in residues at interaction interfaces or buried deep within the protein, and had a more destabilising impact on protein function.
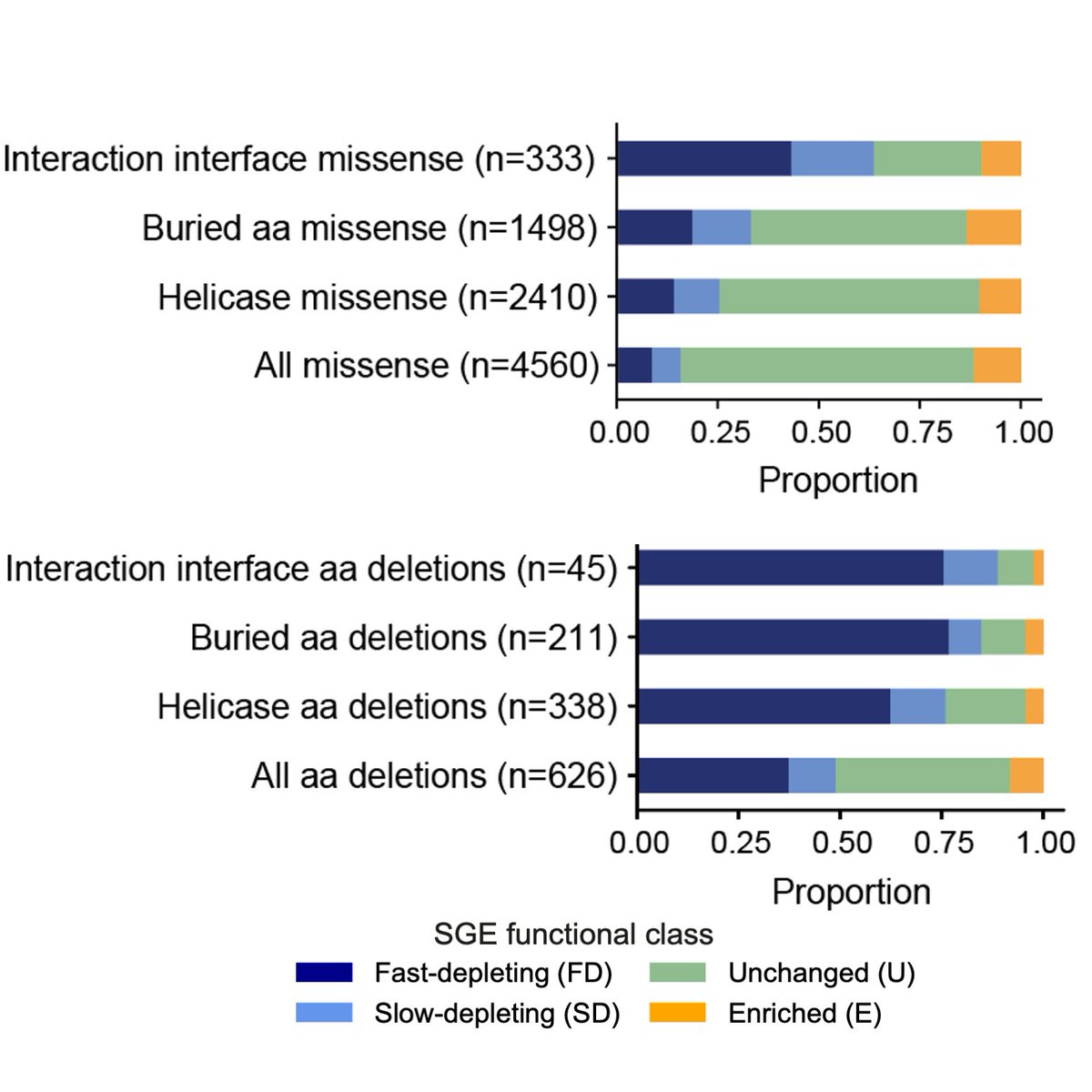

HongKee TAN Matt Hurles @GeretySebastian Greg Findlay Lea Starita Jay Shendure 9. 99.5% of nonsense variants predicted to undergo NMD, 49% of codon deletions and 16% of missense variants were depleted in our assay. 92% of depleted missense variants were in the helicase domains or Q motif. Depleted codon deletions were also highly enriched in these domains.


HongKee TAN Matt Hurles @GeretySebastian Greg Findlay Lea Starita Jay Shendure 10. While only 1% of synonymous variants were functionally abnormal, 23% of canonical splice site variants were functionally normal.

HongKee TAN Matt Hurles @GeretySebastian Greg Findlay Lea Starita Jay Shendure 11. We also identified 105 intronic variants outside of canonical splice sites which were functionally abnormal and one variant in the 5' UTR that generated an upstream, out-of-frame start codon.
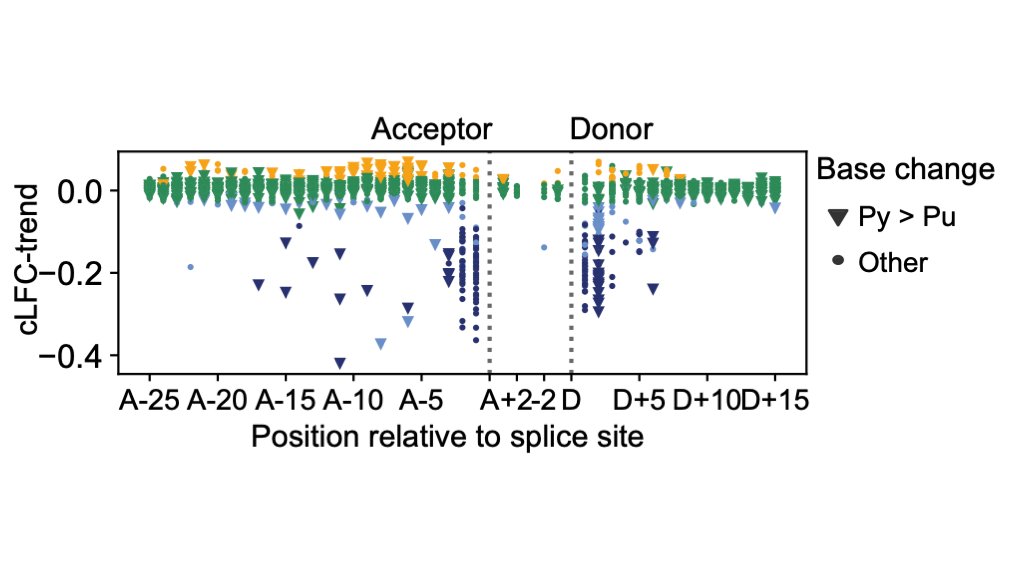

HongKee TAN Matt Hurles @GeretySebastian Greg Findlay Lea Starita Jay Shendure 12. We do not fully understand the enriched variants. They appear to be under purifying selection and are over-represented in conserved missense variants and intronic variants of greater splice disrupting potential, but are not yet associated with any specific phenotype.

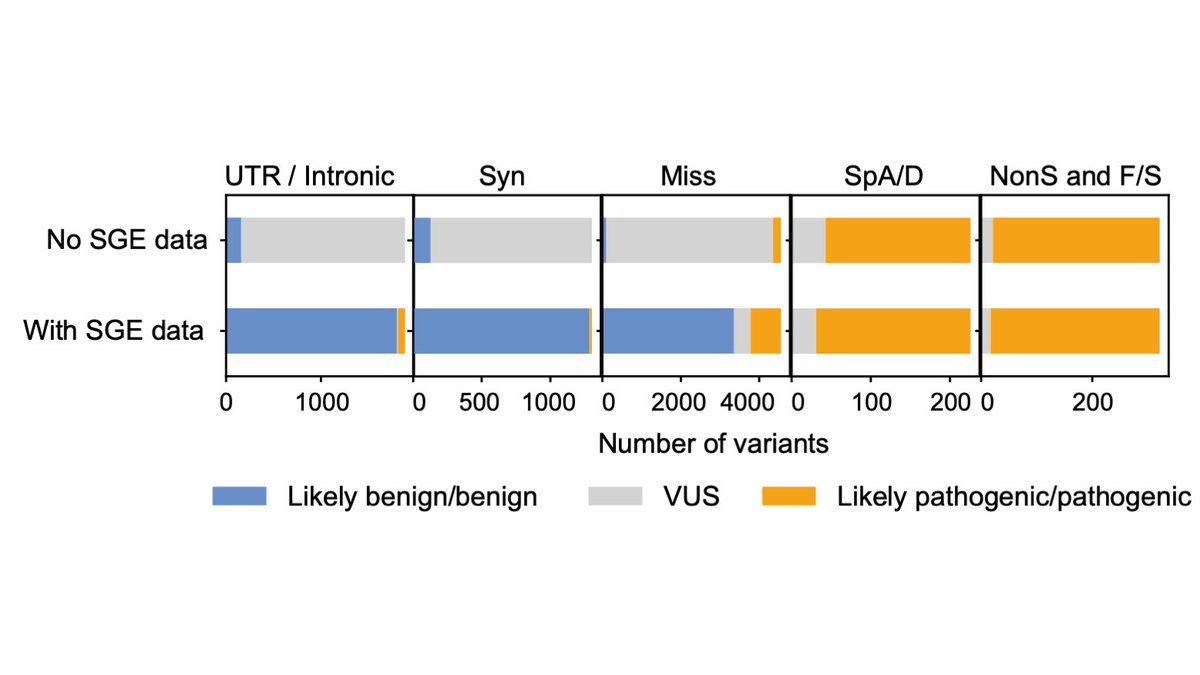
HongKee TAN Matt Hurles @GeretySebastian Greg Findlay Lea Starita Jay Shendure 13. Over 700 DDX3X variants observed in population cohorts and in NDD patients were used to train a random forest classifier to identify functionally abnormal variants of NDD-relevance. This had an accuracy of 99%, substantially outperforming in silico predictors.

HongKee TAN Matt Hurles @GeretySebastian Greg Findlay Lea Starita Jay Shendure 14. Our modelling suggests that using these data in clinical variant classification will reduce the number of VUS in DDX3X by up to 93%. Supplementary data 6 contains predictions of NDD-relevance for all variants, and is intended for use in clinical variant classification.

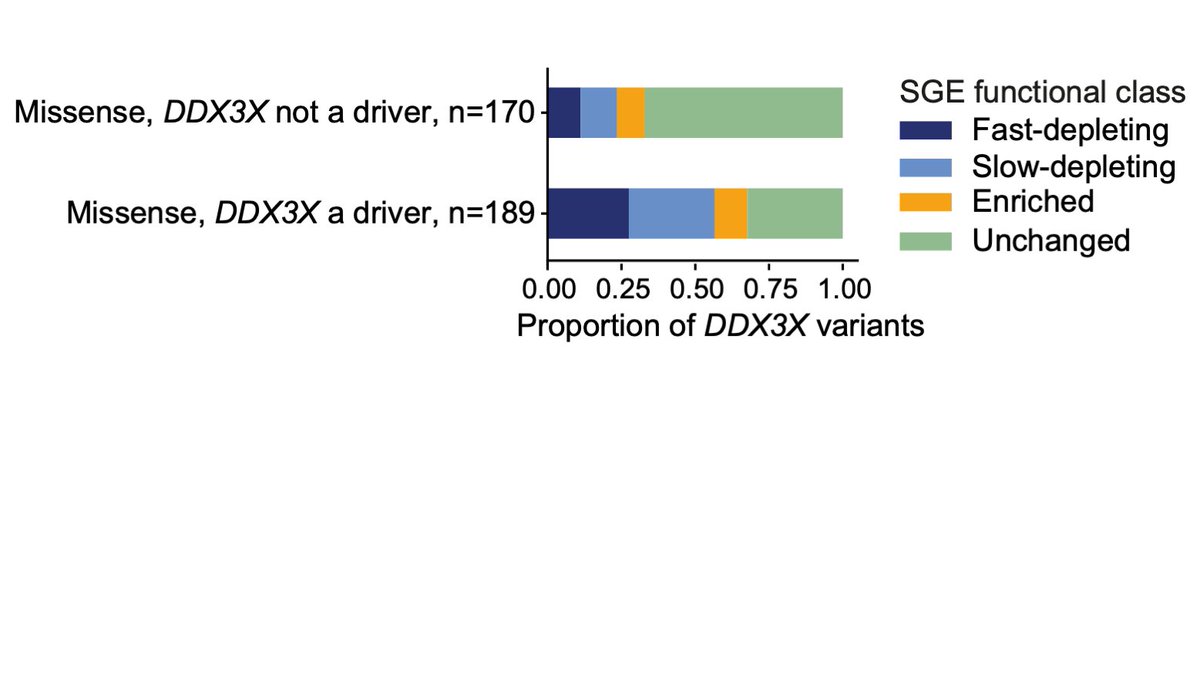
HongKee TAN Matt Hurles @GeretySebastian Greg Findlay Lea Starita Jay Shendure 16. SGE-depleted variants are over-represented in somatic mutations in cancer types where DDX3X is a driver. Depleted variants can account for almost all of the excess of DDX3X mutations in medulloblastoma. Together this suggests that DDX3X functions as a tumour suppressor.

HongKee TAN Matt Hurles @GeretySebastian Greg Findlay Lea Starita Jay Shendure 17. In summary, this map of DDX3X variant effects is highly informative for NDD and cancer, despite being made in a cell type of minimal pathophysiological relevance. This suggests that, for DDX3X, pathogenic variation is protein-intrinsic with minimal cell context dependency.

HongKee TAN Matt Hurles @GeretySebastian Greg Findlay Lea Starita Jay Shendure 18. We are hopeful that use of these data in clinical variant classification will reduce the number of VUS and speed up diagnosis of DDX3X-related neurodevelopmental disorder. Clarifying the role of DDX3X in cancer may guide future therapeutic developments.

HongKee TAN Matt Hurles @GeretySebastian Greg Findlay Lea Starita Jay Shendure 19. We are excited to be applying SGE to other developmental disorder genes. This will be a community effort supported by the Atlas of Variant Effects alliance. Variant.effect.org

HongKee TAN Matt Hurles @GeretySebastian Greg Findlay Lea Starita Jay Shendure On a personal level, this has been a hugely enjoyable project. I have also had two babies since we began experiments, and I am so grateful for the support of my co-conspirator @honkeeT and the mentorship of @SebastianGerety and Matt Hurles. Here’s to the next 100 genes! End of 🧵


Saturation genome editing of DDX3X clarifies pathogenicity of germline and somatic variation Matt Hurles @GeretySebastian pubmed.ncbi.nlm.nih.gov/38057330/

