
David Dylus
@ddylus
ID: 33806718
21-04-2009 04:10:20
138 Tweet
83 Followers
130 Following

Read2Tree: an assembly-free comp. genomics method for small & large-scale projects. Works on @Pacbio Oxford Nanopore Illumina& RNA/DNA & low coverage 0.2x+. Get your phylo tree from raw reads in minutes! e.g #COVID19 biorxiv.org/content/10.110… David Dylus Christophe Dessimoz BCM HGSC #Bioinformatics
Fritz Sedlazeck presents Read2Tree: a new assembly-free approach to directly infer phylogenetic trees efficiently from raw reads with 0.2x coverage+. Come hear how it applied to Coronaviridae samples Room: G With David Dylus, Adrian Altenhoff, Sina Majidian, and Christophe Dessimoz #ISMB2022

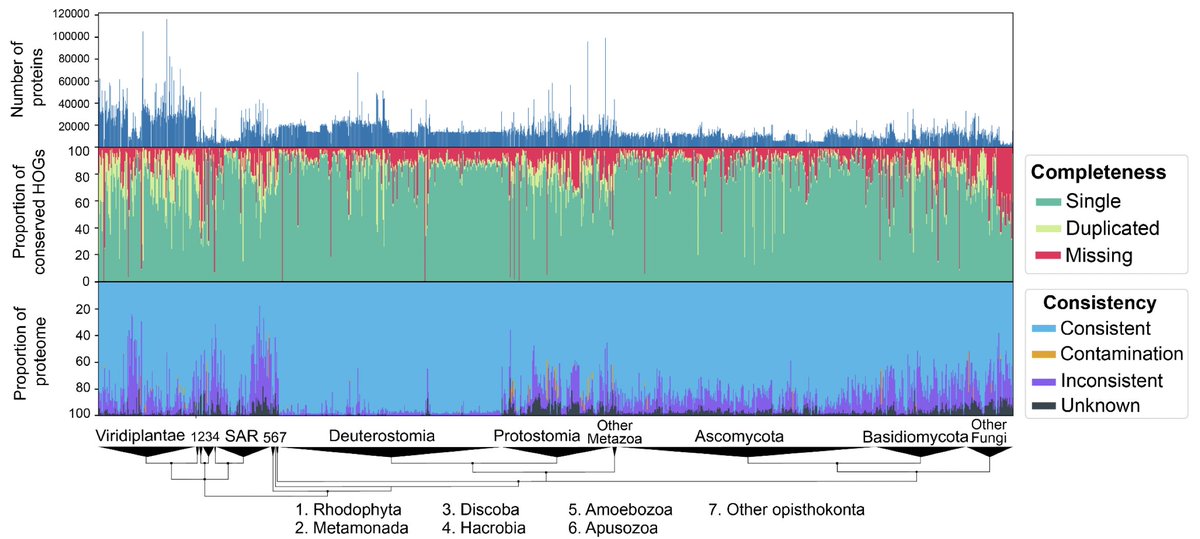
Genome annotation quality affects so many things we do. Our brilliant Yannis Nevers Yannis Nevers devised a tool to assess proteomes—quick and easy. It reports not only *missing* but also *dubious* content! Blog post: lab.dessimoz.org/blog/2022/12/1… Preprint: doi.org/10.1101/2022.1…

Looking for a #postdoc? We're looking for candidates to apply to the Beatriu de Pinos Postdoc program to join our lab Metazoa Phylogenomics Lab Institute of Evolutionary Biology (IBE). 3-year contract and competitive salary. Possibility of funding until publication of selected candidates 1/2

!!!PLEASE RT!!! Postdoctoral fellowship opportunity. Phylogenetics of deuterostomes (and testing their possible non-monophyly). Come and work with the Telford and Yang labs at UCL. jobs.ac.uk/job/CYK582/res…. Ziheng Yang's Group UCL Faculty of Life Sciences CLOE: UCL Centre for Life's Origins and Evolution UCL Biosciences !!!PLEASE RT!!!


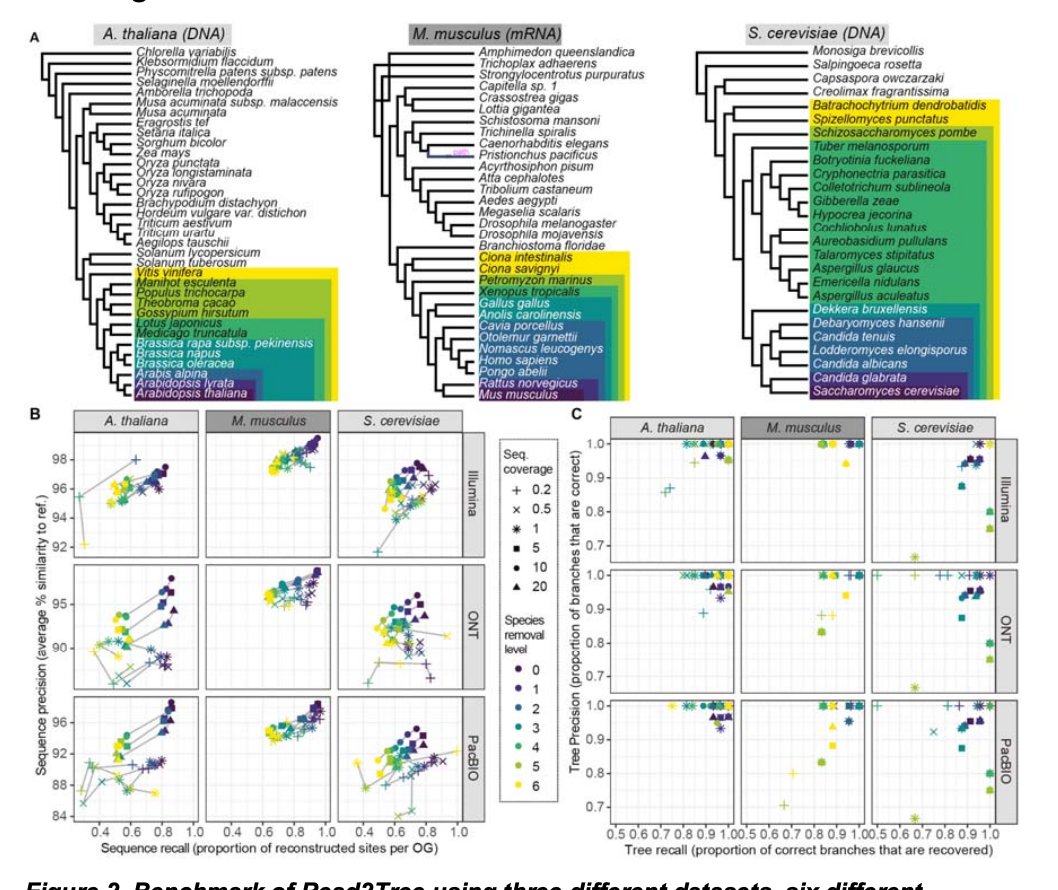
Super happy that the Read2Tree paper is now out at Nature Biotechnology—the centrepiece of David Dylus’s postdoc and a wonderful collaboration with Fritz Sedlazeck. Key contributions by Sina Majidian and Adrian Altenhoff too. nature.com/articles/s4158…

Université de Lausanne SIB ETH Zurich BCM HGSC Rice Computer Science UCL Computer Science CLOE: UCL Centre for Life's Origins and Evolution Fritz Sedlazeck David Dylus Co-lead authors Christophe Dessimoz and Fritz Sedlazeck go #behindthepaper on a method to build phylogenetic trees directly from raw reads, bypassing time-consuming steps such as genome assembly. Plus: what Read2Tree has in common with the song "Smoke on the Water" go.nature.com/3KAZcz0

Bioinformatics can address these challenges! At SIB, we have researchers on it (e.g. Daniele Silvestro Uni Fribourg & Rob Waterhouse also @rmwaterhouse.bsky.social Université de Lausanne), an environmental working group, and we are now looking for a *Director for Environmental Bioinformatics*. Apply or share! apply.refline.ch/499599/0280/pu…

This is part of an exciting Swiss National Science Foundation -funded interdisciplinary grant with 𝘋𝘪𝘳𝘬, Iñaki Ruiz-Trillo and Dave Moi. This position is to develop cutting edge methods for evolutionary structural biology. Please apply or help us spread the word!

The length distribution of proteins is remarkably uniform across the tree of life ➡️ universal selection force at play. Paper by postdoc extraordinaire Yannis Nevers, with Lecompte & Natasha Glover. 📝Blog post + backstory lab.dessimoz.org/blog/2023/06/1… 📰Paper doi.org/10.1186/s13059…

Reminder that we have 2 postdoc openings: 1) position on synteny and gene loss in 🍅w/ @SebastianSoyk x.com/Gloveface/stat… 2) evolutionary structural biology to reconstruct the proteome & interactome of the first animal (urmetazoa) w/ Pedro Beltrao x.com/pedrobeltrao/s…

Pls RT: 4 weeks until the registration deadline of our #Bioinformatics #Science #Hackathon BCM HGSC. Come join us & work on exciting prototypes across DNAnexus, Inc. PacBio Oxford Nanopore GREGoR Consortium SMaHT Network. Prizes for the best teams! Aug30-Sept1: hgsc.bcm.edu/events/hackath…


Could you envision a future where inferring #orthology for thousands of species becomes possible?! The future is already here and I'll be presenting FastOMA in #ISMBECCB2023 in Lyon! See you in EvolCompGen EvolCompGen on Thursday 27th at 15:30! Try it out github.com/DessimozLab/Fa…



Did you catch Natasha Glover's talk about bringing science to the public at #SMBE2023? Be sure to also check out her GBE article: Homoeolog Inference Methods Requiring Bidirectional Best Hits or Synteny Miss Many Pairs buff.ly/44K6aJN

Happy to see a summary about Read2Tree from our Nature Biotechnology paper from my home country Austria - Österreich (German only sorry): science.apa.at/power-search/1… Link to paper nature.com/articles/s4158… Christophe Dessimoz Sina Majidian David Dylus BCM HGSC Rice Computer Science

Here a fantastic piece! Three years of major work (lead by Elise Parey and coordinated by Ferdinand Marlétaz). Including the genome, an analysis of bilaterian chromosome evolution and a comparison of global expression patterns in bilaterian appendage regeneration. biorxiv.org/content/10.110…

So happy to see our brittle star genome led by 𝗘𝗹𝗶𝘀𝗲 𝗣𝗮𝗿𝗲𝘆 finally published in NatureEcoEvo highlighting the evolution of echinoderm chromosomes as well as the origin of the regeneration programm nature.com/articles/s4155…

