The DECIPHER Project
@deciphergenomic
ID: 4501798349
https://www.deciphergenomics.org 16-12-2015 10:11:03
444 Tweet
2,2K Followers
104 Following

Estimated population penetrance for variants associated with cardiomyopathies are now displayed alongside cardiac allele frequencies. This information is useful when considering secondary findings. Kathryn McGurk James Ware and @cvgenomics | #cardiogen
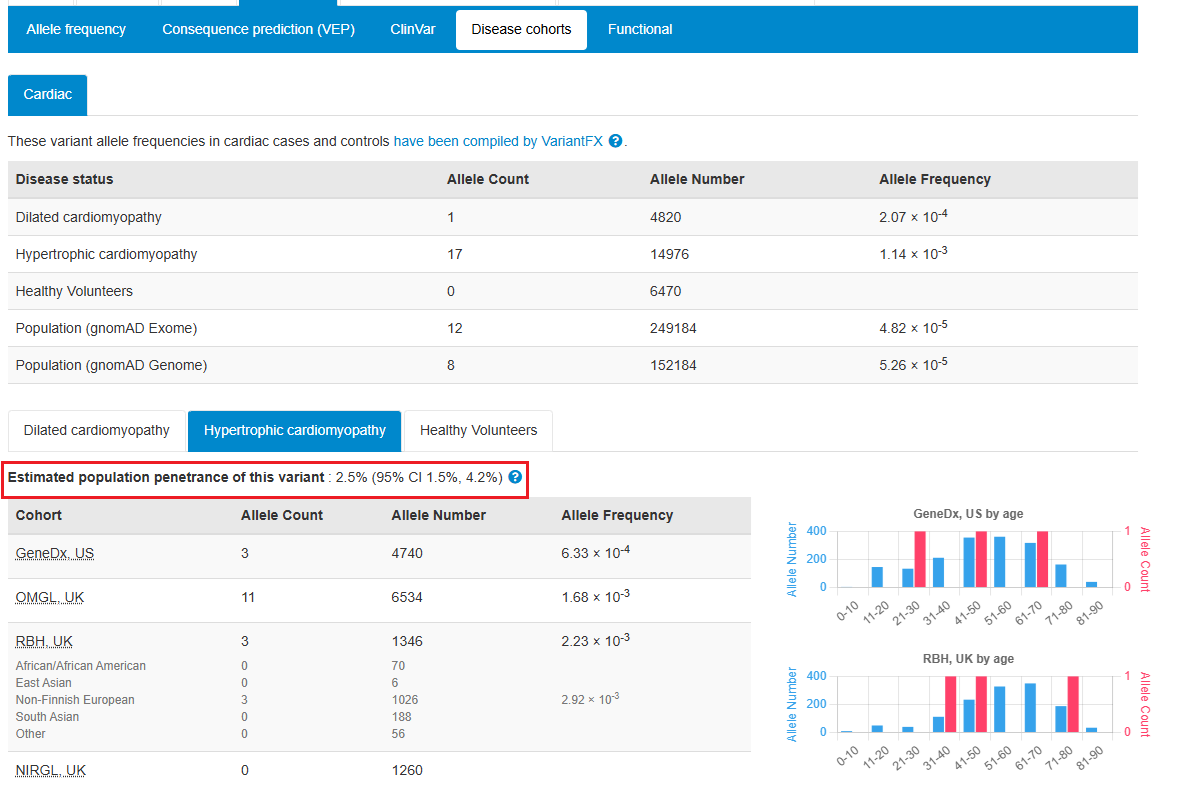

Cardiac case/control cohort data, which demonstrates the confidence of cardiac gene-phenotype relationships associated with specific variant classes, has been updated; more variant classes e.g. canonical splice site variants Kathryn McGurk James Ware and @cvgenomics #cardiogen


The prevalence for cardiomyopathies is also now displayed for genes associated with cardiac disorders Kathryn McGurk James Ware and @cvgenomics | #cardiogen



A Genome Aggregation Database Short Tandem Repeat track is now available on the genome browser which displays information about 60 disease associated repeat loci. The associated diseases are displayed along with the normal and pathogenic repeat lengths, and links to STRipy


DECIPHER users can now search for patient matches in Boston Children's seqr in addition to PhenomeCentral, Broad Institute seqr, GeneMatcher, RD-Connect and MyGene2 using MatchmakerExchange












Human Developmental Cell Atlas (HDCA) expression data is now displayed. Expression is displayed in 12 sections of a 6-7 post-conception week human embryo, alongside a sagittal view which displays the region of the embryo represented by each section Haniffa Lab Muzlifah Haniffa