Diego
@deigsmnns
Something similar to a Biologist and a Bioinformatician. PhD student at CNIC.
ID: 1017383440126181376
12-07-2018 12:21:28
43 Tweet
97 Followers
723 Following

Diego @MTorrescnic David Sancho CNIC CNIC BIoIT Hopefully many people will be using our new tool, as they already did with the original digitalDLSorter, pioneer work using deep learning on scRNASeq data for deconvolution on bulk rnaseq frontiersin.org/journals/genet… We keep on working to improve their performance! 💪🏻💪🏻💪🏻





What a PERFFect day to share our new preprint! With Tsion Abay #BobStickels Meril Takizawa @ChaligneRonan and Ansu Satpathy, we introduce PERFF-seq, a new experimental approach to studying rare cells with scRNA-seq via transcript-specific enrichment. biorxiv.org/content/10.110… 1/n

Álvaro García from the MechanoCaveoLab together with Diego Mañanes from David Sancho kick off the #AtheroConvergence HR20 CaixaResearch closure meeting in beautiful Donostia CIC biomaGUNE CNIC YaleVBTProgram Yale University Aarhus Universitet



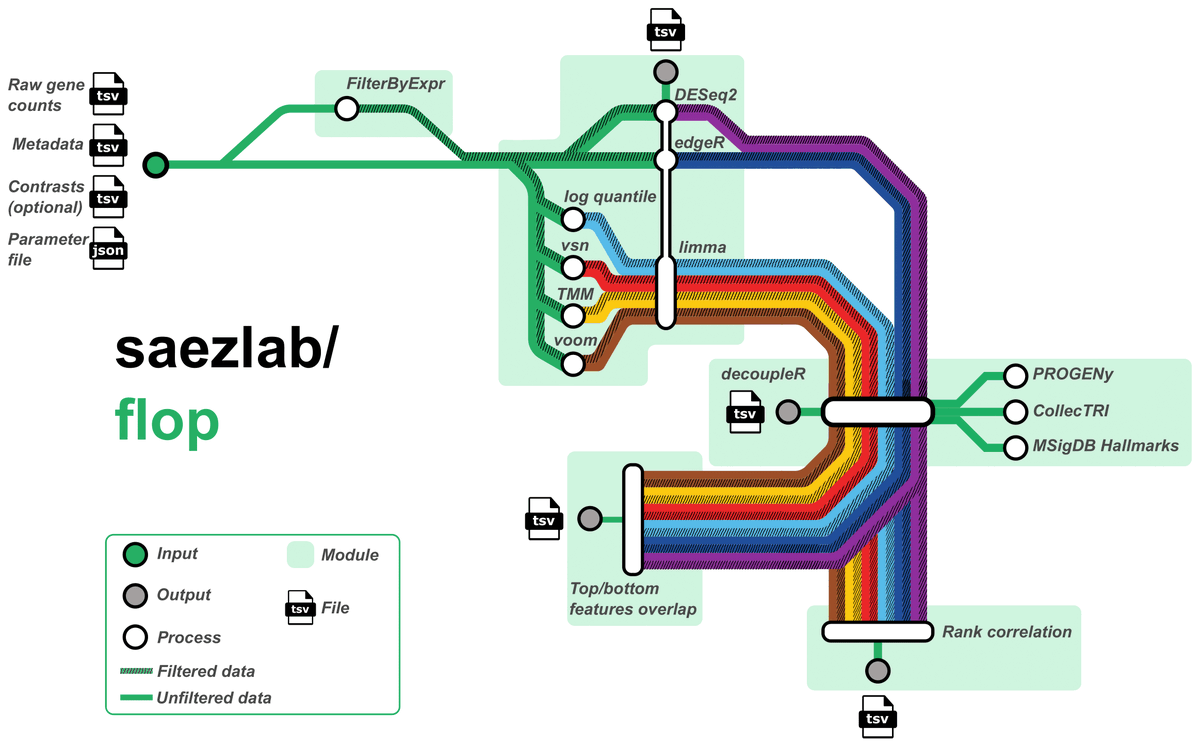
Our workflow, FLOP, exploring the impact of filtering, normalization, and differential expression on functional analysis is now published in Nucleic Acids Res. doi.org/10.1093/nar/gk…