DingquanYu
@dingquan_yu
PhD candidate in computational bio | Whatever torch we kindle, whatever space it may illuminate, our horizon will always remain encircled by the depth of night
ID: 1555669109861400578
https://github.com/dingquanyu 05-08-2022 21:36:54
59 Tweet
42 Followers
55 Following





A new version of AlphaPulldown by DingquanYu. Includes experimental support for multimeric templates in AlphaFold by Dmitry Molodenskiy and interface to crosslink-based modeling AlphaLink2 of Rappsilber laboratory github.com/KosinskiLab/Al…

Work is in full swing on AlphaPulldown by DingquanYu and Dmitry Molodenskiy, but with so many users, thousands of downloads, and 34 open issues, incl. many exciting feature requests, help would be welcome! If you want to contribute, feel free to fork and submit a pull request!


The final version of the PyTME paper published in SoftwareX Journal! PyTME enables fast CPU and GPU-based template matching in #cryoET and fitting in #cryoEM. More in the thread with plenty of screenshots👇 sciencedirect.com/science/articl…


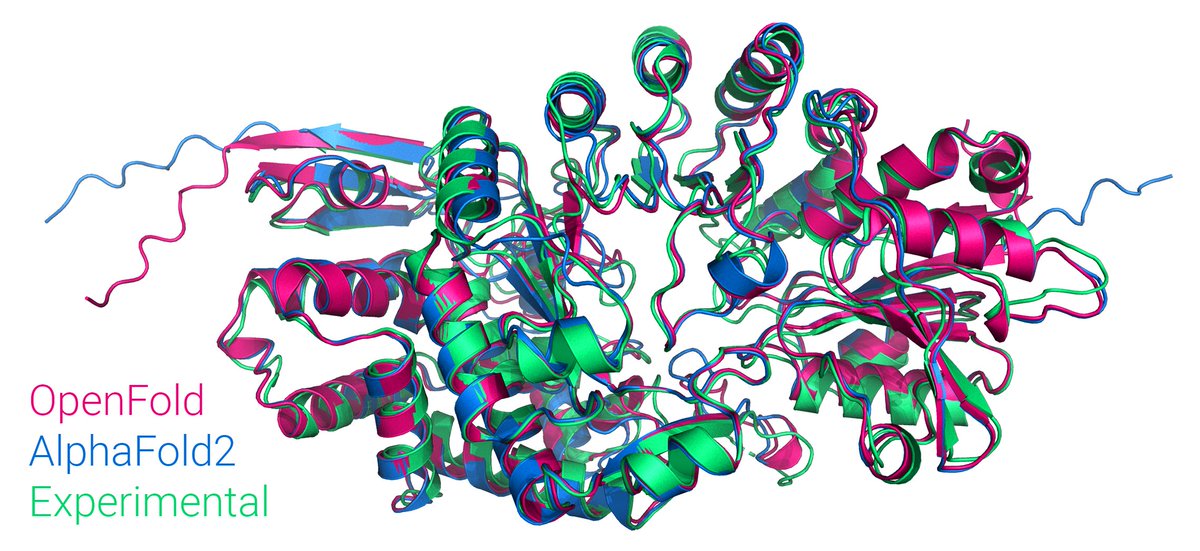
I’m greatly thankful to Jan Kosinski and Mohammed AlQuraishi for this opportunity. The openfold team has provided me with invaluable help and inspirations. Excited for the upcoming developments in OpenFold!





OpenFold paper is now out in Nature Methods. Timely. I’ll say more about AF3 at the end of this thread, but first I wanted to highlight what’s new since our initial release. (1/9)

bioRxiv Bioinfo Thanks Alexandre Bonvin - also on Bluesky for testing this and citing and using AlphaPulldown! In AlphaPulldown you can also limit to one model by selecting one model name and using it as the input for the --model_names flag. We can try to emphasize this in the new manual we’re working on, but maybe

Two 🔥 computational postdoc positions 🔥are available in our lab, funded by European Research Council (ERC) ! embl.org/jobs/position/… and embl.org/jobs/position/…. Join us and unleash the power of these just-delivered beasts 👇And please repost to help us!