Divya Koyyalagunta
@divyakoyy
PhD student in Computational Biology & Medicine @ Tri-I (@WeillCornell/@MSKCancerCenter). Previous software engineer @Apple health and cs @DukeU. she/her/hers
ID: 1458136849420017667
09-11-2021 18:18:44
46 Tweet
158 Followers
155 Following


So honored to receive this award! A million thanks and congratulations to my partner in crime Julia Zhao, and to ISCB and ISMB organizers for an all-around amazing conference! One of my favorites to date🎉

So excited to be in Hawaii ICML Conference presenting work w/ Quaid Morris, and to win best poster award!! Thanks for a great workshop ICML-Compbio-Workshop


Thank you to all organizers and sponsors CZI Science GSK for another outstanding ICML-Compbio-Workshop workshop ICML Conference #ICML2023 photos courtesy of Lingting Shi



Super excited to share the first preprint from Karuna Ganesh Lab! An amazing collaboration with dana_peer, led by the phenomenal andrew moorman @f_cambuli and Ellie Benitez Memorial Sloan Kettering Cancer Center. Progressive #Plasticity during #ColorectalCancer #Metastasis. biorxiv.org/content/10.110…

Very grateful to receive the U.S. National Science Foundation grfp! And a huge thank you to Quaid Morris and Tri-I CBM for the support!


✨New paper alert Cancer Discovery✨ - GDD-ENS distinguishes 38 cancer types (>96% of patients) w/ high accuracy using routinely collected DNA sequencing data #MSKIMPACT and generalizes well to other gene panels w/o retraining - CNAs (arm/gene level) are key features for classification


Metient reconstructs the migration histories of metastatic cancers by scoring a Pareto front of parsimonious histories. It can be auto-calibrated to different cohorts. Superstar PhD student Divya Koyyalagunta will present at #ISMB2024 in MLCSB Check it out: biorxiv.org/content/10.110…




Thrilled to share our paper now out nature delineating #plasticity during #CRC #metastasis, a fantastic collaboration led by Andrew Moorman Ellie Benitez, and @f_cambuli from the Karuna Ganesh Lab and @dana_per lab! nature.com/articles/s4158… rdcu.be/dYAVK. A 🧵





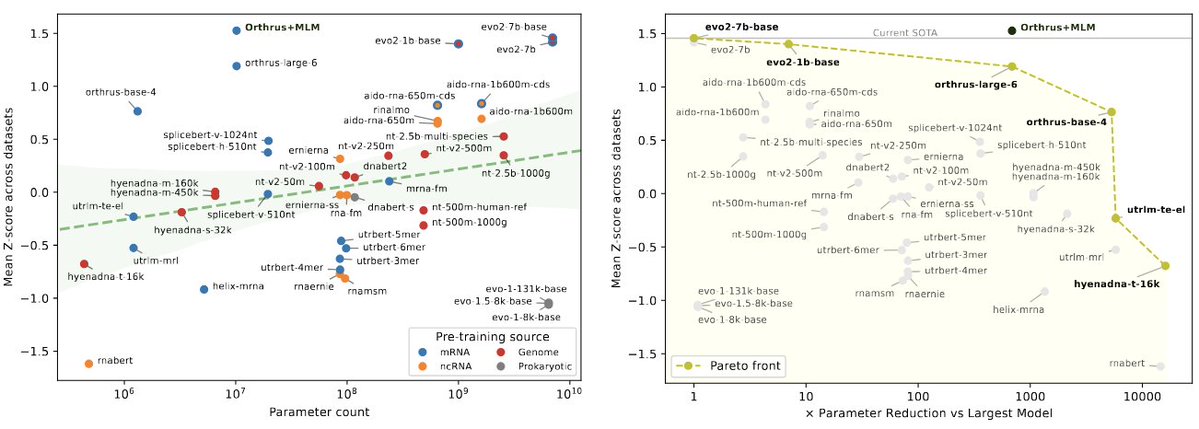
(4) Together with Divya Koyyalagunta, we further assess the ability for foundation models to compositionally generalize from learned motifs. Models are exposed to either sequence element that promotes translation, but never both, and we task them with predicting the unseen combination.