
Eugenio F. Fornasiero
@euforna
Interested in aging, molecular turnover, neurons, synapses, super-resolution microscopy, metabolic labeling and proteomics
ID: 2202313684
https://www.fornasierolab.uni-goettingen.de/ 30-11-2013 21:43:51
540 Tweet
521 Followers
454 Following

🚀The Milan #RNASalon is on! 21 labs working on RNA biology from 10 institutions in Milan meeting monthly for scientific talks, new collaborations, networking and much more. Kick-off 29 Nov at HT. Many thanks to The RNA Society and Lexogen for their support! humantechnopole.it/en/trainings/m…



I am very happy to share our work revealing new aspects of the cell biology of neurons with the dendritic axon origin. It was a great pleasure to collaborate with Christophe Leterrier and his team! rupress.org/jcb/article/22…


Gene regulation involves thousands of proteins that bind DNA, yet comprehensively mapping these is challenging. Our paper in Nature Genetics describes ChIP-DIP, a method for genome-wide mapping of hundreds of DNA-protein interactions in a single experiment. nature.com/articles/s4158…

Happy to announce the 8th Schram Foundation Symposium in Göttingen on the 25ht of March 2025 (satellite of the German Neuroscience society). Attendance is free and we have great speakers including Tomohisa Toda, Marlene Bartos Pawel Burkhardt Schaefer Lab Please circulate!



As we know, the human plasma🩸 proteome is crucial! The Matthias Mann Lab just published a fantastic study in Nature Genetics! And here is our News & Views highlighting the mapping of plasma protein QTLs and why #MassSpec remains a key player: nature.com/articles/s4158…. Am Soc for Mass Spec Human Proteome







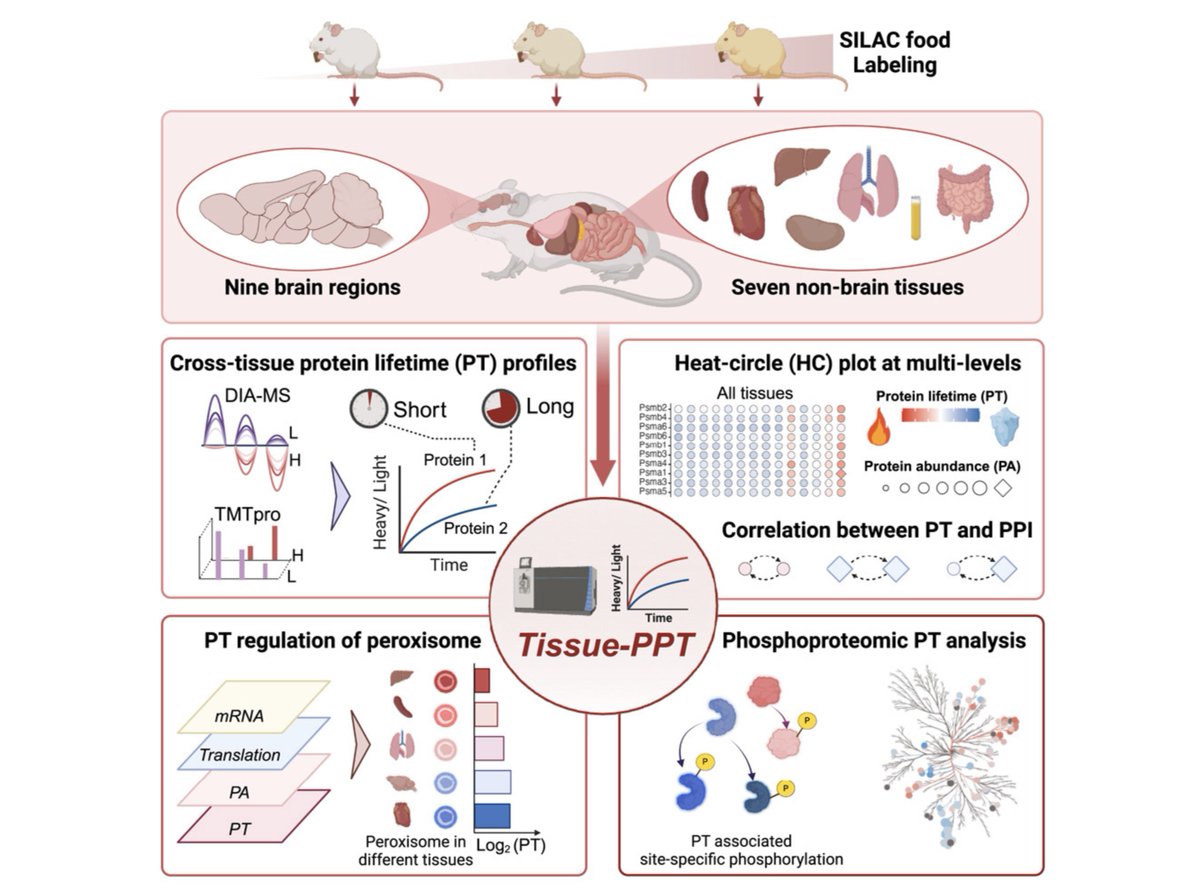
6/ Explore our data through Turnover-PPT 🔗[yslproteomics.shinyapps.io/tissuePPT] This work would not have been possible without our collaborators in Junmin Peng Eugenio F. Fornasiero @shisheng2017🤝🤝, the hard work of our lab members Andy, and supportive environment at Yale University Yale West Campus.
![Yansheng Liu (@yanshen73854711) on Twitter photo 6/ Explore our data through Turnover-PPT 🔗[yslproteomics.shinyapps.io/tissuePPT]
This work would not have been possible without our collaborators in <a href="/JunminPengLab/">Junmin Peng</a> <a href="/euforna/">Eugenio F. Fornasiero</a> @shisheng2017🤝🤝, the hard work of our lab members <a href="/Andy01230396/">Andy</a>, and supportive environment at <a href="/Yale/">Yale University</a> <a href="/YaleWestCampus/">Yale West Campus</a>. 6/ Explore our data through Turnover-PPT 🔗[yslproteomics.shinyapps.io/tissuePPT]
This work would not have been possible without our collaborators in <a href="/JunminPengLab/">Junmin Peng</a> <a href="/euforna/">Eugenio F. Fornasiero</a> @shisheng2017🤝🤝, the hard work of our lab members <a href="/Andy01230396/">Andy</a>, and supportive environment at <a href="/Yale/">Yale University</a> <a href="/YaleWestCampus/">Yale West Campus</a>.](https://pbs.twimg.com/media/GmfumWSaQAAtdq1.jpg)

🚀 Excited to share our latest work! Proud to have been part of this collaborative work on Protein turnover across several tissues and brain regions Cell with Yansheng Liu and Junmin Peng #proteomics authors.elsevier.com/c/1koBJL7PXuE9g

🚀 Excited to share our latest work! Proud to have been part of this collaborative work on Protein turnover across several tissues and brain regions Cell with Yansheng Liu and Junmin Peng and @NishaHemandha from the lab. Check it out👇 authors.elsevier.com/c/1koBJL7PXuE9g

A pair of methods papers for studying synapses: DELTA: a method for brain-wide measurement of synaptic protein turnover reveals localized plasticity during learning Boaz Mohar (@[email protected]) karel svoboda @rhodamine110 Nelson Spruston Eugenio F. Fornasiero HHMI | Janelia Allen Institute nature.com/articles/s4159…







