
Charley Yen
@eugeniecyen
@London_NERC_DTP PhD student in @EizaguirreLab applying genomics and epigenomics for sea turtle conservation 🌊🐢🧬 previously @CamZoology @stjohnscam🦋 she/her
ID: 1288970004889772032
30-07-2020 22:49:39
303 Tweet
316 Followers
491 Following



We found a record of the overlapping generations of F108 in Dept of Zoology while replacing the door label today Ana Pinharanda Joe Hanly Kathy Darragh Gabriela Montejo-Kovacevich 🇪🇺 Anna Orteu Charley Yen Eva van der Heijden Dheeraj Halali and soon-to-be Dr Rachel Blow


Fun to present our newer collaborative results on sex determination in #seaturtle using #DNAMethylation part of Charley Yen s PhD project. In short, can we use DNAmeth for sexing? Yes! Natural Environment Research Council QM_SBBS Queen Mary University of London



Featuring my concentration face whilst loading precious sea turtle DNA samples into the Oxford Nanopore machine, for our 1st time sequencing genomes in the field! 😆 #WorldTurtleDay

Ending the day with Charley Yen stunning the room showing her work to sex #seaturtles using #DNAMethylation. We hope this work will guide further management for the protection of those endangered species globally. #ECCB2024


Charley's talk was also a hit! 🥳 It was fantastic to see how engaged people were about our work developing epigenomic tools to monitor sublethal thermal stress for #seaturtle #conservation 🐢🥚🌡️#SMBE2024 Charley Yen Project Biodiversity


Excited to share our new paper out in Nature Communications! We used experimental evolution to understand how gene body methylation might evolve and participate in adaptive evolution. Great team effort with @PSarkies, Jennifer Westoby & Rebecca Kilner! a 🧵: rdcu.be/dPY94

Happy to share our preprint by Charley Yen et al! We produced a chromosome-scale genome & methylome for a loggerhead #SeaTurtle from the Cabo Verde nesting group 🇨🇻🐢 We use these resources to yield #genomic & #epigenomic insights for #conservation of these threatened species


Charley Yen Project Biodiversity @alice_balard QM_SBBS Martín-Durán lab Rossiter Lab @ QMUL We match past population size changes to major climatic events 🌍 highlight microchromosomes as special regions of genetic and epigenetic diversity 🧬 & create a functional association & methylation map from 191 genes linked to temperature-dependent sex determination 🌡️♀️♂️


Happy to share our paper is now out in Evolutionary Applications ! We identified over 200 differentially methylated sites that could be used to monitor thermal stress beyond mortality in endangered #SeaTurtles 🐢 doi.org/10.1111/eva.70…


Excited to present at the #PAG2025 Oxford Nanopore workshop tomorrow! Come along to hear more about how we incorporate ONT methylation sequencing for sea turtle conservation 🐢🧬

There were remarkable updates presented at #PAG32 on incredibly accurate Oxford Nanopore sequencing for applications ranging from telomere-to-telomere plant & animal genome assemblies, to amplicon sequencing at massive scale. Here are some highlights in case you missed them (thread)
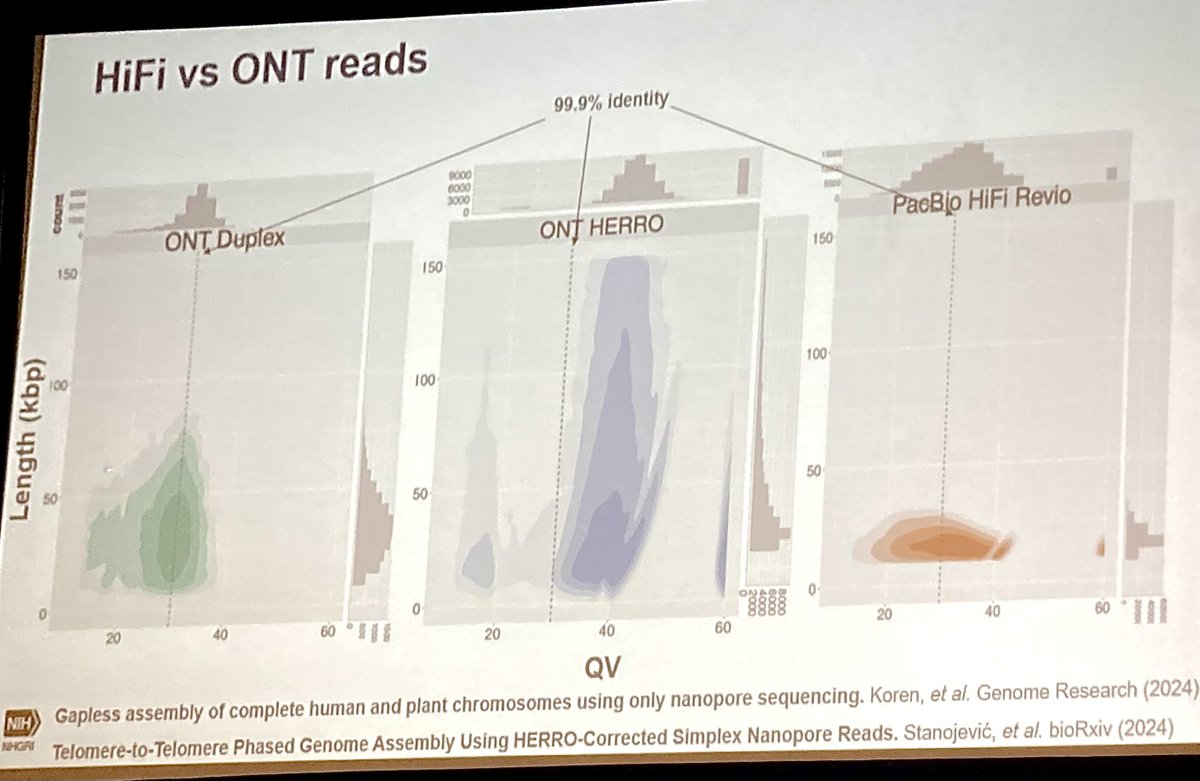

Eugenie 'Charley' Yen (Queen Mary U of London) then shared her beautiful work using Oxford Nanopore for genomic & epigenomic approaches for conservation of endangered loggerhead sea turtles - even taking the P2 device into the field to deploy methylation-based biomarkers in real-time!
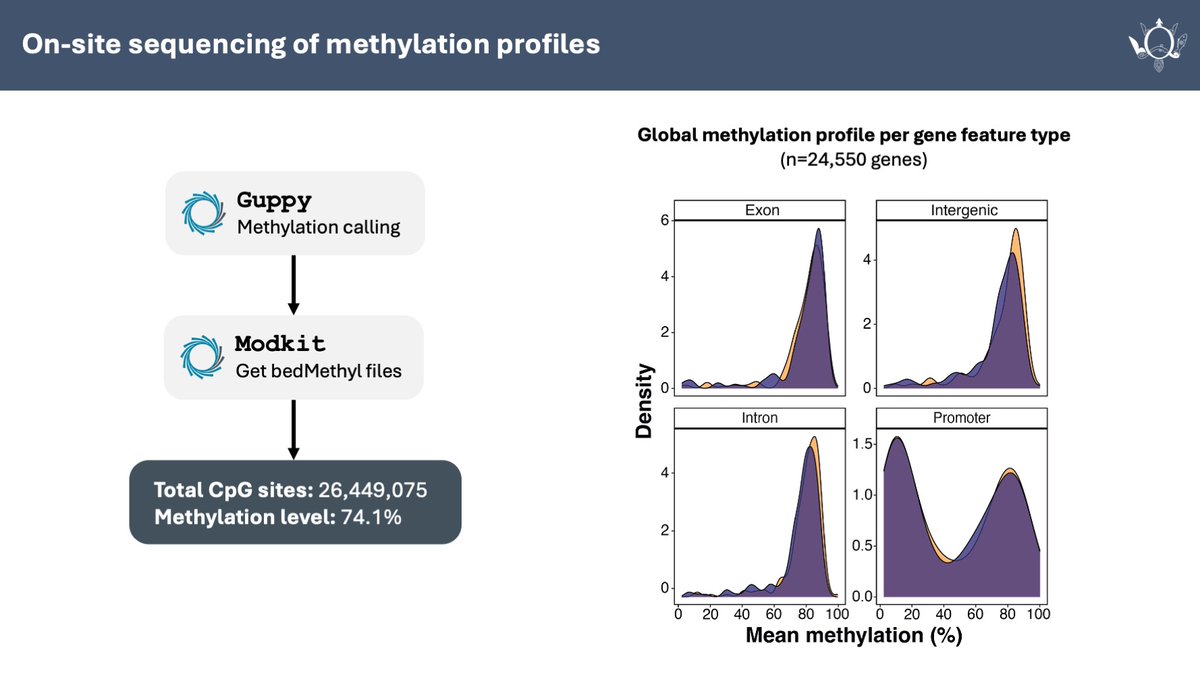

Turtley cool usage of Oxford Nanopore sequencing to simultaneously generate genomic and epigenomic resources simultaneously. Chromosome-level genome assembly and methylome profile yield insights for the conservation of endangered loggerhead sea turtles doi.org/10.1093/gigasc…



