Fabian Theis
@fabian_theis
Computational biologist @HelmholtzMunich, prof @TU_Muenchen & associate PI @sangerinstitute. Dad of 4 and mountain lover. Department news, see @CompHealthMuc
ID: 121690511
https://www.helmholtz-munich.de/en/icb/pi/fabian-theis 10-03-2010 08:25:56
4,4K Tweet
33,33K Followers
427 Following

🏹 Job alert: The Institute of Computational Biology at Helmholtz Munich | @HelmholtzMunich seeks a Postdoc Researcher (f/m/x) in ML or Computational Biology to join the Theis Lab w/ Oxford Centre of Neuroinflammation. 📍 Neuherberg near Munich 📅 Deadline: Apr 11 🔗 bit.ly/3Rlu1KD

How can causal machine learning be applied to single-cell genomics to understand the true biological mechanisms underlying cellular processes?Nature Genetics TU München "Causal machine learning for single-cell genomics" • Advances in single-cell '-omics' technologies combined


Great joint work with Alejandro Tejada Lapuerta Paul Bertin Stefan Bauer Yoshua Bengio Fabian Theis. Link to the paper: nature.com/articles/s4158…


New nature Insightful perspective on large language of life models for molecular and cell biology nature.com/articles/s4158… Bo Wang Fabian Theis Haotian Cui and colleagues



Thrilled to share our Human Endoderm-derived Organoid Cell Atlas (HEOCA). We compare organoids with primary tissues, assess culture strategies, and explore disease models. In collaboration with Lennard Halle, MD, supervised by Gray Camp, Fabian Theis & Treutlein lab.Institute of Human Biology



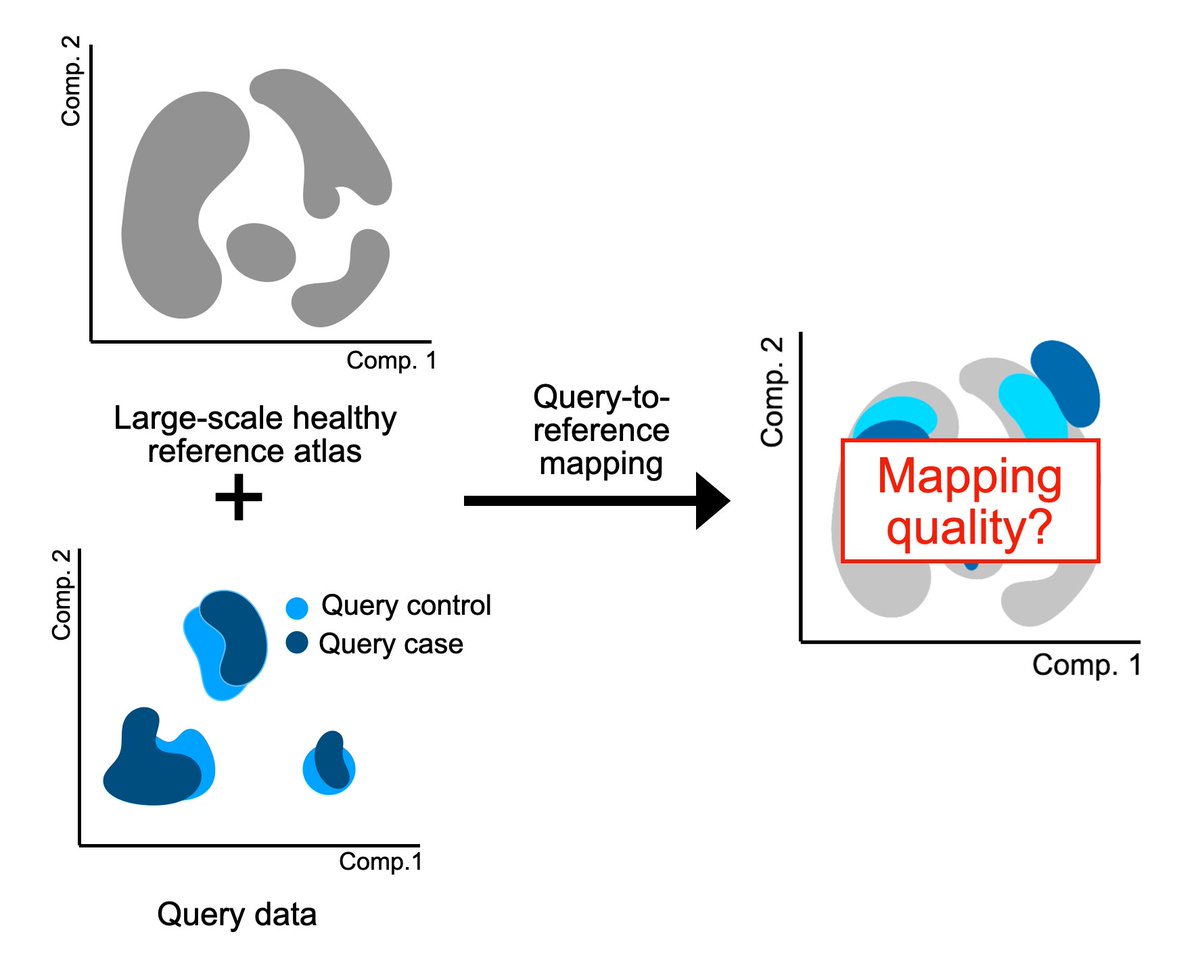
Analyzing your single-cell data by mapping to a reference atlas? Then how do you know the mapping actually worked, and you’re not analyzing mapping-induced artifacts? We developed mapQC, a mapping evaluation tool biorxiv.org/content/10.110… from the Fabian Theis lab. Let’s dive in🧵