Ferhat Ay
@ferhatay
dad x 2, husband, son, brother, computational biologist, genome scientist, associate professor at la jolla institute lji.org
ID: 89387775
https://www.lji.org/labs/ay/ 12-11-2009 06:16:42
1,1K Tweet
1,1K Followers
1,1K Following

Fantastic work from our friend and collaborator Karine Le Roch at UC Riverside. Culmination of her many years of work to fight against malaria! 👏👏


It was a 4days of great scientific discussion, brainstorming and some fun along the way! Big thanks to U.S. National Science Foundation for funding & NSF-NCEMS leaders & organizers for a well-organized summit! All that was left for us was to turn coffee into ideas and proto-proposals :) #NCEMSSummit










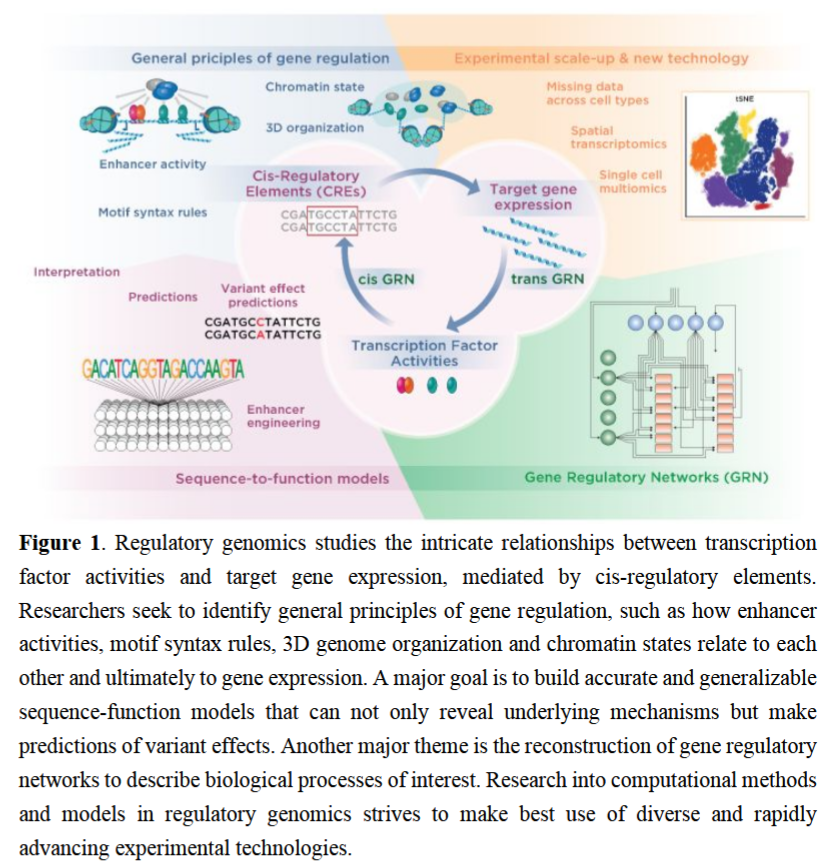
Perspective on recent developments and challenges in regulatory and systems genomics academic.oup.com/bioinformatics… is out, amazing to think about this with Julia Zeitlinger (@[email protected]), Ferhat Ay, Shaun Mahony, Anthony Mathelier, Alejandra Medina-Rivera, Jason Ernst!
🧬 Just out in Bioinformatics Advances: "Perspective on recent developments and challenges in regulatory and systems genomics" Find the full paper here: doi.org/10.1093/bioadv… Authors include: Julia Zeitlinger (@[email protected]), Sushmita Roy @sroyyors.bsky.social,genomic.social, Ferhat Ay, Anthony Mathelier, Alejandra Medina-Rivera, Shaun Mahony,