Francesca Grisoni
@fra_grisoni
Associate Prof. #AI for drug discovery | Leading @molecularML at @TUeindhoven & @ICMStue | @ELLISforEurope Scholar | Previously @ETH & @unimib | she/her 🏳️🌈
ID: 954373517306400768
19-01-2018 15:22:32
791 Tweet
4,4K Followers
934 Following

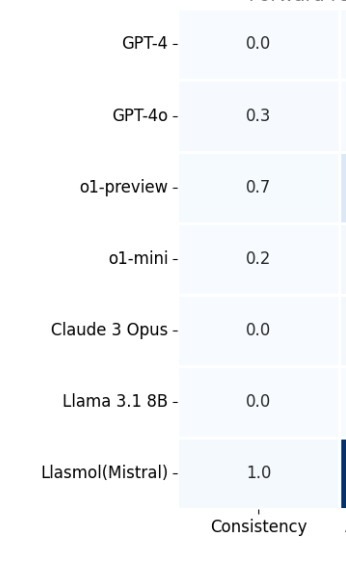
How can #MachineLearning & #automation accelerate the understanding and design of complex molecular systems? Have a look at our latest preprint, an interdisciplinary collaboration between 3 teams van Hest Lab Molecular Machine Learning & Brunsveld’s lab💪🏻 Read Andrea Gardin post to know more👇
ELLIS ML4Science workshop co-organised by our PI Francesca Grisoni & https://bsky.app/profile/aidd.bsky.social PI Günter Klambauer was a great event to learn advances in ML & promote https://bsky.app/profile/aidd.bsky.social results: ai-dd.eu/lectures. https://bsky.app/profile/aichemist.bsky.social fellows Dina Khasanova Marcel Hiltscher attend poster session. Berlin eve.


Thanks for the visit and the inspiring talk on #AI for antibiotic discovery, César de la Fuente ! Fascinating research at the intersection of biology, chemistry and machine learning 💪🏻 Huge thanks to Antoni Forner-Cuenca for co-organizing the event and to TU/e Artificial Intelligence Systems Institute for its support!


Great news to conclude 2024! I couldn’t think of a better team than the ones and only narf42 and Robert Pollice to work on this exciting project. 🚀 Cool times ahead for the collaboration between TU Eindhoven and University of Groningen!



Happy that our “Hitchhiker’s guide” to the universe of chemical language processing is out Digital Discovery. 🚀 👽 We provide guidelines on how to use deep learning to learn the “chemical language” of bioactivity. 🧪 Spearheaded by the one and only Rıza Özçelik 💪🏻 Molecular Machine Learning



Are you using generative #DeepLearning for de novo molecule design?🧪 🖥️ Then check out Rıza Özçelik ‘s latest work, where we perform a (super) large scale analysis (~1 B designs!) & find ‘traps’, ‘treasures’ and ‘ways out’ in the jungle of generative drug discovery. 🌴 🐒 👇

"The Jungle of Generative Drug Discovery: Traps, Treasures, and Ways Out" by Rıza Özçelik , Francesca Grisoni Paper: arxiv.org/abs/2501.05457 #machinelearning


Proud of Helena Brinkmann’s first PhD paper on rethinking how SMILES augmentation is performed for generative #DeepLearning! 🚀 Check it out! 👇 📄 chemrxiv.org/engage/chemrxi… 🖥️ github.com/molML/fantasti… Molecular Machine Learning TU Eindhoven


📢📢📢The ML-GUIDE team (Francesca Grisoni, Robert Pollice and myself) are looking for 3 PhD students, passionate about combining the directed evolution of diverse biomolecules with deep learning approaches to develop better (bio)catalysts and drugs! Pls retweet tinyurl.com/3jz97ter

🚨Open PhD positions TU Eindhoven & University of Groningen! Are you intrigued by the potential of ML+directed evolution in drug discovery (Francesca Grisoni), peptide catalysis (Robert Pollice) or biocatalysis (narf42)? Do you like interdisciplinary, cutting-edge research? Then apply!👇

We just preprinted a fresh study on molecular machine learning on OOD molecules 🧠 Using joint modeling, we could detect distribution shifts, estimate prediction reliability, and capture meaningful molecular patterns! doi.org/10.26434/chemr… #AI #chemistry Molecular Machine Learning