
santosh
@funtoshs
Biochemist. Transitioning from studying protein conformation in a tube @JNCASR to cells @TheCrick with @balchin_david using mass spectrometry.
ID: 2368785104
02-03-2014 12:08:42
272 Tweet
105 Followers
197 Following

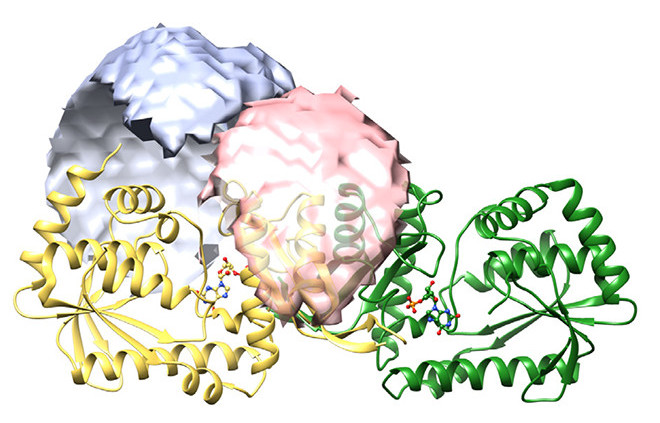
NH3 channeling enzymes typically form tight complexes, but the M jannaschii GMP synthetase subunits interact transiently. Balaram and colleagues JNCASR Bangalore now present a mechanistic model explaining the various steps in this fascinating catalytic cycle. pubs.acs.org/doi/full/10.10…

I’m very proud to share the first paper from my lab The Francis Crick Institute, a wonderful collaboration with John R. Engen. Made possible by Aleksandra Pajak and Thomas Wales, with contributions from santosh and Alzbeta Roeselova. A summary🧵: 1/11 biorxiv.org/content/10.110…

3yr #postdocposition in Cambridge AstraZeneca, split between their shiny new DISC & our lab Babraham Institute. Help explore how cellular stresses shape #PROTAC efficacy, & vice-versa. Fundamental/translational relevance aplenty! Oh, some cool #proteomics too. careers.astrazeneca.com/job/cambridge/…

Registration is open for the 33rd annual meeting of the UK Chaperone Club on 17 April 2023! Sessions will cover topics including the structure and folding of molecular chaperones and cellular stress responses. Proteintech Group | New England Biolabs crick.ac.uk/whats-on/uk-ch…

Words of advice: Teaching macromolecular crystallography European Crystallographic Association (ECA) ACAxtal BCA Education BCA CNC_Crystals rcsb pdb 💉🧬💻🔬💊🌱🧠🦠 Protein Data Bank Magnus Kjaergaard ASBMB IUCr Biochemical Society Biophysical Society Crystallography World ccp4_mx Plz RT for greater outreach #XRAY #crystallography

Join us for a PhD! I am recruiting a student to explore uncharted space in protein biogenesis, together with an exceptional team in a fantastic research environment The Francis Crick Institute. Fully funded and open to any nationality crick.ac.uk/careers-study/…

Proud to share our work on molecular chaperones, previously posted as bioRxiv, now out in Molecular Cell David Balchin Sarah Maslen santosh Grant Pellowe J. Mark Skehel The Francis Crick Institute sciencedirect.com/science/articl…


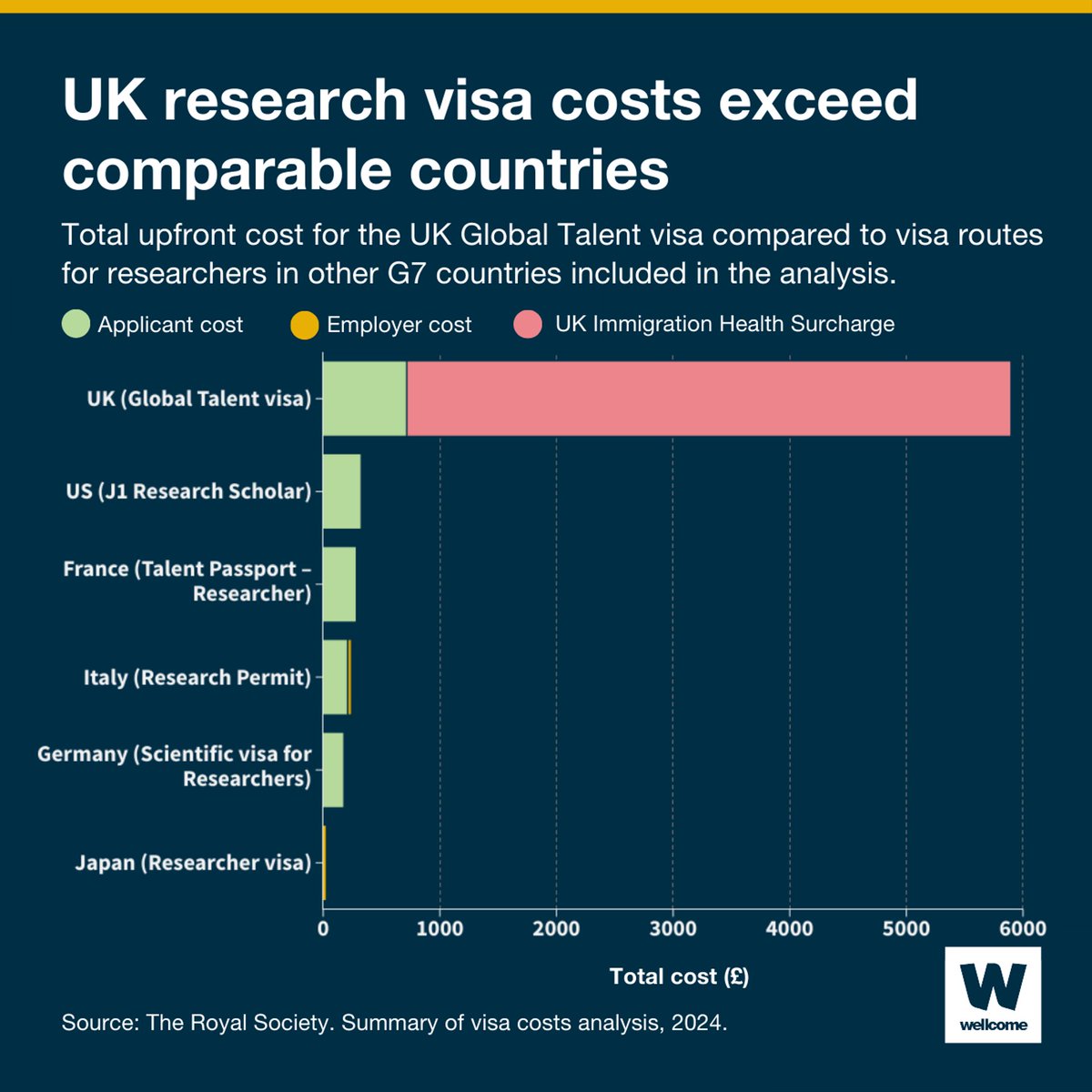
New analysis from the The Royal Society shows just how high UK visa costs for researchers are compared to upfront costs for similar visa routes in 13 other countries. Here, we take a look at how the UK compares to countries in the G7 included in the analysis 🔍⤵️ 1/5



Our #FragPipe-Analyst manuscript is out! pubs.acs.org/doi/10.1021/ac…. Load FragPipe's TMT/ LFQ/DIA quant tables (peptide or protein quant) and get QC, Limma, PCA, volcano plots, heatmaps, do GO/pathway enrichment analysis, all with just a few clicks! Tutorials:fragpipe-analyst-doc.nesvilab.org

New from our team The Francis Crick Institute, led by Grant Pellowe. Eukaryotes are very good at folding multidomain proteins. Here we give some insight into how the human ribosome helps with this, by tuning how nascent domains fold and dock. 1/n biorxiv.org/content/10.110…



Starting with 2 ug of protein for S-trap digestion, I end up with 15 ug of tryptic peptides as reported by Thermo BCA colorimetric peptide assay. Where am I going wrong? #TeamMassSpec Bini Ramachandran Brett Phinney @ProteomicsNews