
Gabriele Scalia
@gabo_scalia
ID: 816947113280077824
05-01-2017 09:59:04
10 Tweet
40 Followers
26 Following




Excited to share Tangram, our method for mapping scRNAseq data on spatial data. Our work is part of the #BICCN and #HubMAP consortia. Collaboration led by Aviv Regev, between Broad Institute, Harvard University, AI Skunkworks at Northeastern University and FattahAmin X Unifi. biorxiv.org/content/10.110…
We are proud to share our paper with a comprehensive single cell RNA-seq, ATAC-seq and spatial atlas of the developing mouse neocortex, led by Daniela Di Bella from the Arlotta Lab and Ehsan Habibi from Aviv Regev’s lab. nature.com/articles/s4158…

In particular we now map all clusters at E11.5 to a full 3D embryo, with help of the excellent Tangram algorithm by Tommaso Biancalani and Gabriele Scalia of Regev lab (github.com/broadinstitute… with our tweaks at github.com/linnarsson-lab…)
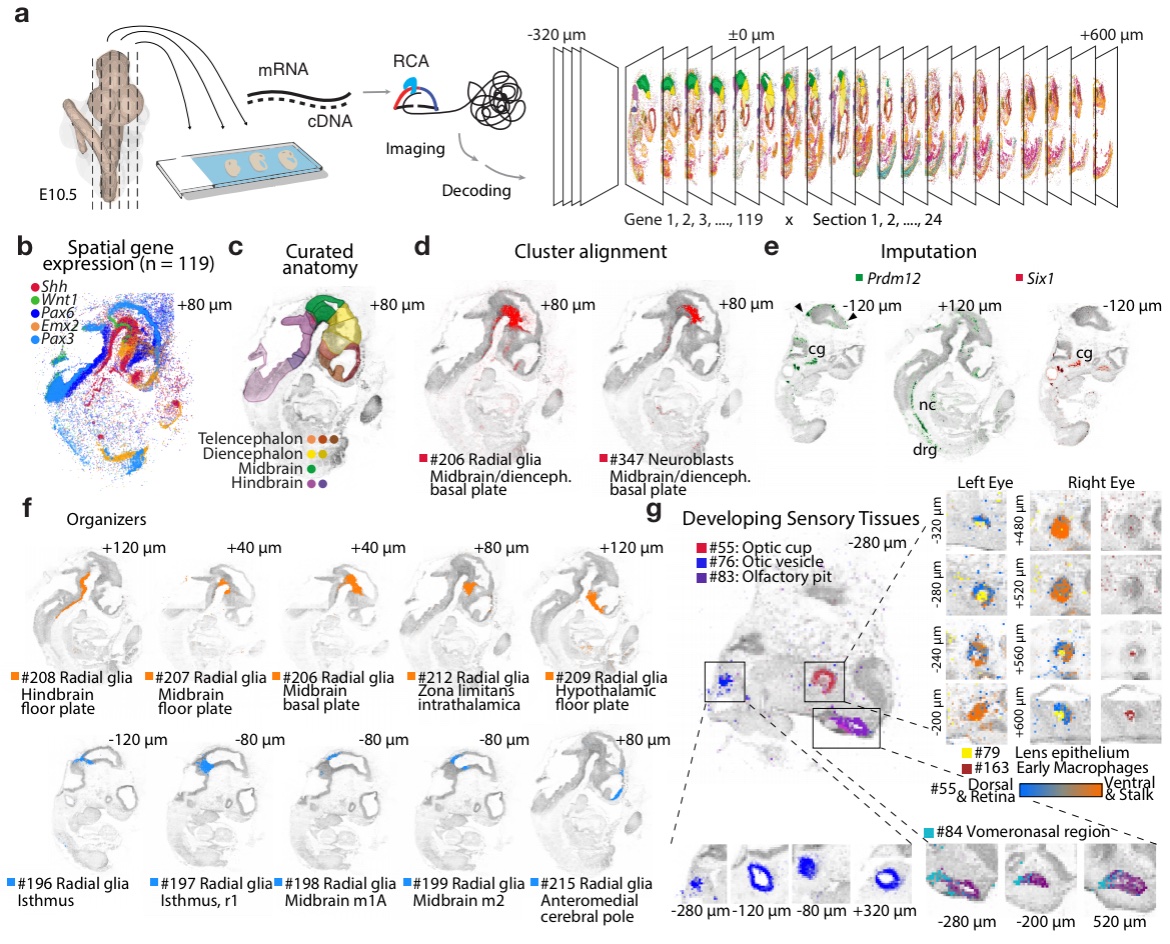

Tangram is a versatile tool for aligning single-cell and single-nucleus RNA-seq data to spatially-resolved transcriptomics data using deep learning. Broad Institute Tommaso Biancalani Gabriele Scalia OA paper: nature.com/articles/s4159…


Why should diffusion models be linear? With multiple classes, they can be hierarchical! My work with Gabriele Scalia shows that hierarchical diffusion models improves their efficiency _and_ interpretability! arxiv.org/abs/2212.10777






