Vincent Gardeux
@gardeux_vincent
Senior Scientist
ID: 246186099
02-02-2011 09:34:54
89 Tweet
74 Followers
185 Following

Exchange at #SingleCell genomics symposium with researchers from EPFL Université de Lausanne FMI science

A new paper in eLife - the journal by David Suter shows how by #pluripotency transcription factors dynamically regulate #chromatin accessibility across the cell cycle doi.org/10.7554/eLife.…

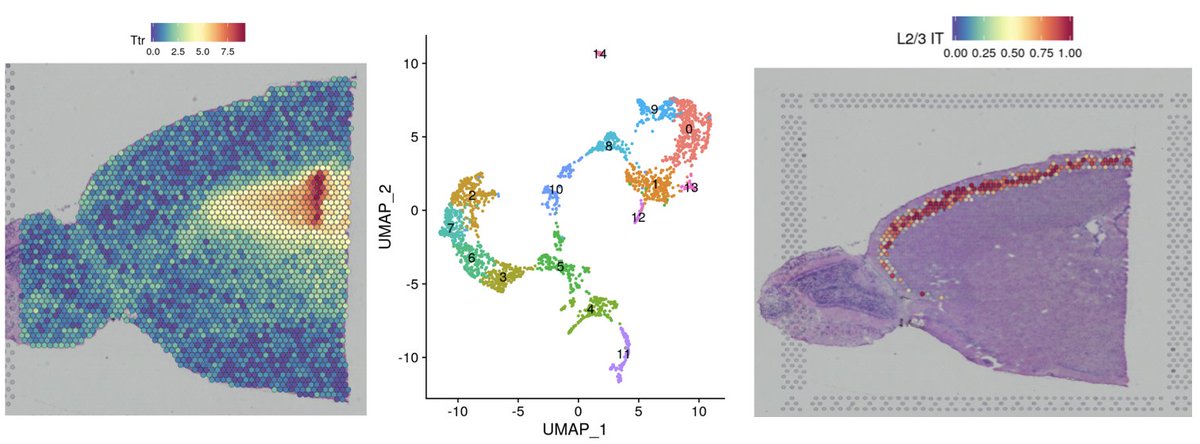
We’re excited to release a Seurat update with support for Spatial Transcriptomics data! Includes clustering, interactive visualization, and integration with scRNA-seq references. Check out our vignette on 10x Genomics Visium data: satijalab.org/seurat/v3.1/sp…

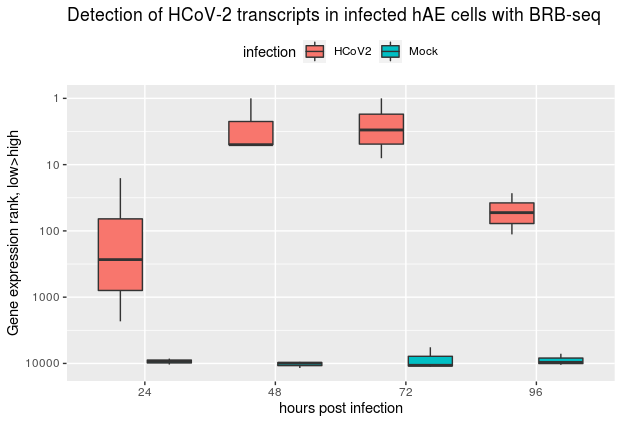
Our exciting work together with Ronald Dijkman shows that we can reliably detect #HCoV2 viral transcripts in infected human cells using BRB-seq (tinyurl.com/y3l5hut8) even at the early stage. Affordable #transcriptomics opens a great perspective for mass #COVID19 diagnostics

Proud of this huge collaborative effort by @PhilipVkovski, Mitra Silvio Steiner, Dr. Jenna Kelly 👩🏻🔬👩🏻💻🦠🧬🇨🇦🇨🇭 Volker Thiel Daniel Alpern, and all other coauthors. Inspired by earlier work by Prof. Akiko Iwasaki. Universität Bern HONOURs-ITN #virology #COVID19 disq.us/t/3o8cdvi

EPFL Professor Andrea Ablasser from EPFL Life Sciences shares the Leenaards Foundation 2020 Science Prize with Professor Michel Gilliet from the CHUV / Centre hospitalier universitaire vaudois. Their project aims to gain insight into the causes and effects of an overactive innate immune system. actu.epfl.ch/news/andrea-ab…






No time to get bored. Half-time retreat Day#3 with Vincent Gardeux Vincent Gardeux EPFL Maira Ihsan EMBL-EBI and Judith Zaugg Judith Zaugg EMBL Marie Skłodowska-Curie Actions
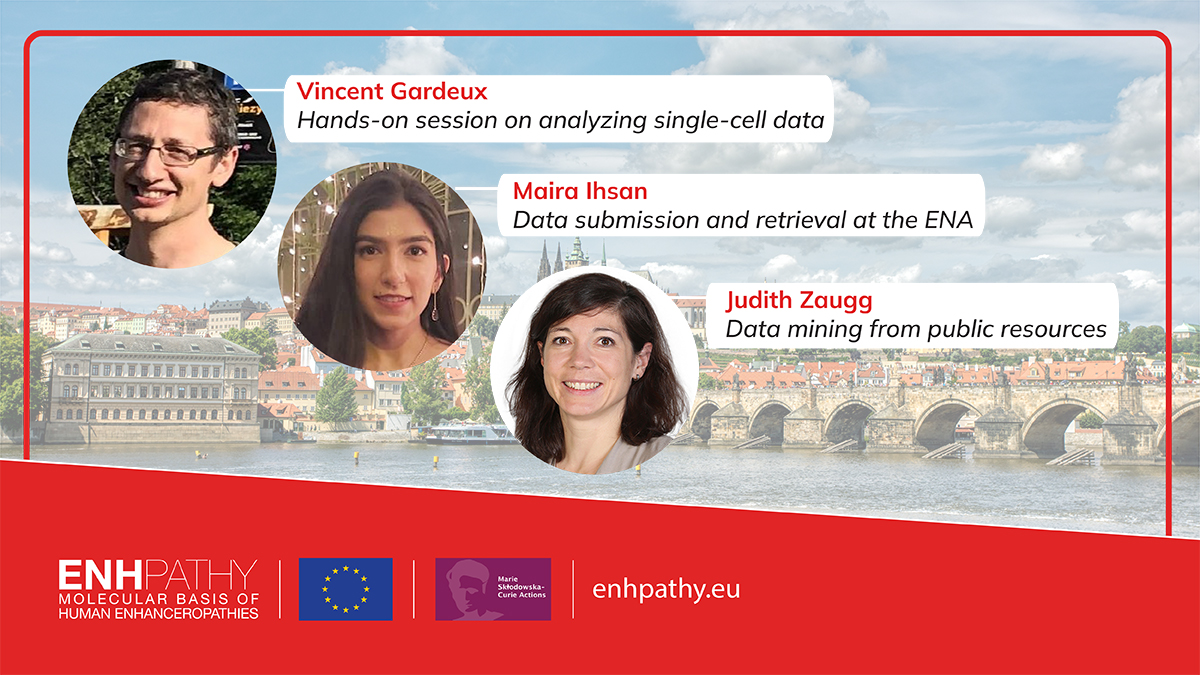

Half-time retreat Day#3 (students' perspective) with Vincent Gardeux EPFL "Hands-on session on analyzing single-cell data" Marie Skłodowska-Curie Actions













![Bgee database (@bgeedb) on Twitter photo A few days left to register with early bird rate to our <a href="/BC2Conference/">[BC]2</a> <a href="/ISBSIB/">SIB</a> workshop "Standardization of single-cell metadata: an Open Research Data initiative" with Jason Hilton of <a href="/cellxgene/">cellxgene</a> & David Osumi-Sutherland <a href="/emblebi/">EMBL-EBI</a> #SingleCell #FAIRdata bc2.ch/tutorials-work… A few days left to register with early bird rate to our <a href="/BC2Conference/">[BC]2</a> <a href="/ISBSIB/">SIB</a> workshop "Standardization of single-cell metadata: an Open Research Data initiative" with Jason Hilton of <a href="/cellxgene/">cellxgene</a> & David Osumi-Sutherland <a href="/emblebi/">EMBL-EBI</a> #SingleCell #FAIRdata bc2.ch/tutorials-work…](https://pbs.twimg.com/media/FzOWxdYWcAAxn3-.png)