Glennis Logsdon
@glennis_logsdon
Assistant Professor at @PennGenetics and @PennEpiInst | T2T, HPRC, & HGSVC member | Loves genomics, epigenomics, & synbio | glennislogsdon.bsky.social
ID: 946258580927377408
http://logsdonlab.com 28-12-2017 05:56:41
499 Tweet
1,1K Followers
334 Following

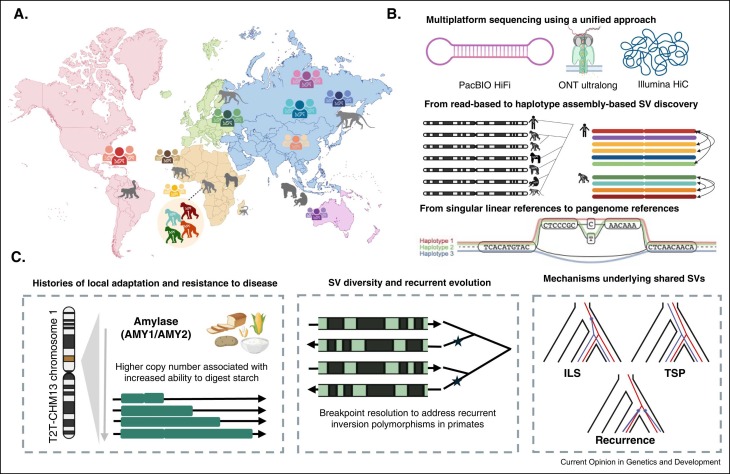
Had so much fun writing this small review with Nicolas Lou and Peter Sudmant about structural variation in primates in the era of T2T genomes and pangenomics :) sciencedirect.com/science/articl…

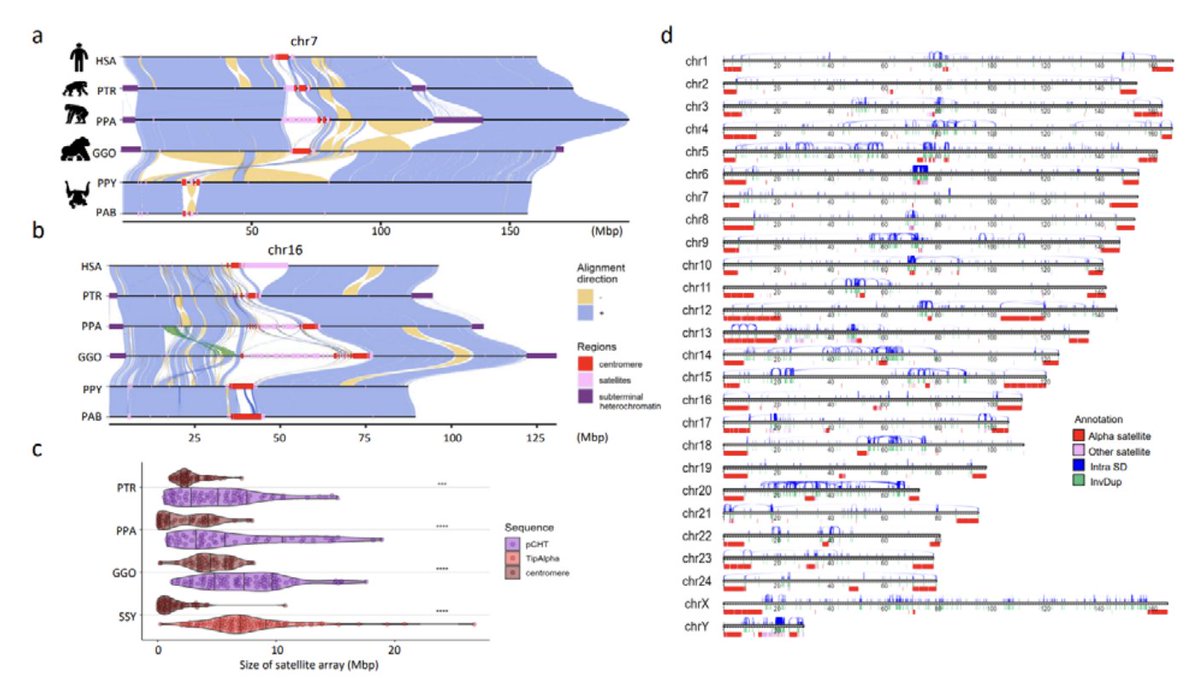
looking forward to this paper on 6 great ape genomes, "great" in both the traditional sense and for assembly quality. From Adam Phillippy EichlerLab Arang Rhie et al; just finished the first 3 pages which means I'm at the end of the author list! 🤣 biorxiv.org/content/10.110…



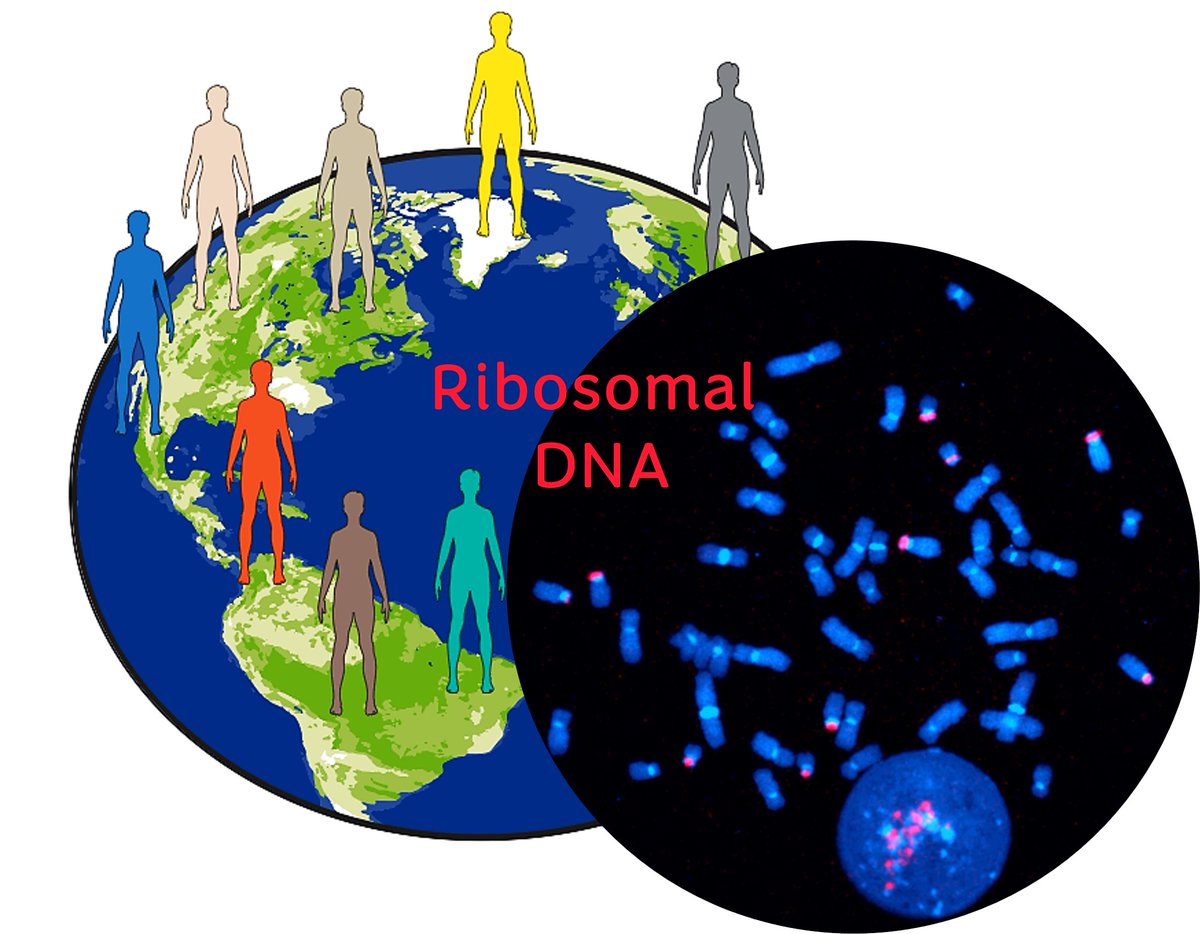
New preprint from our lab 🚨 We explored the variability in ribosomal DNA copy number and activity across humans of the world with Jennifer Gerton Paxton Kostos Andrea Guarracino Steven Solar Erik Garrison Adam Phillippy and others not on X biorxiv.org/content/10.110… 🧵









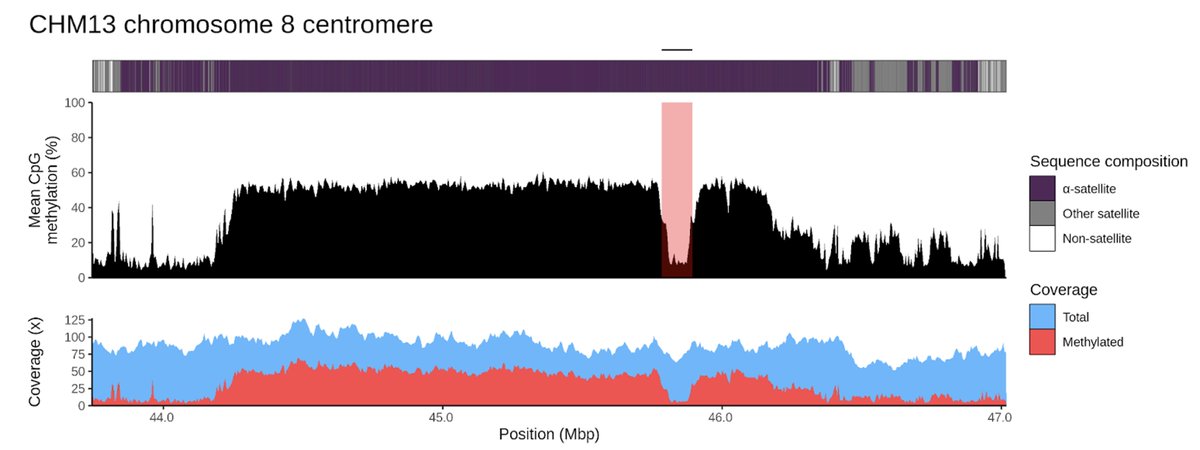
Computational analysis of DNA methylation from long-read sequencing. #DNA #Methylation #LongRead #Sequencing #Genomics Nature Reviews Genetics nature.com/articles/s4157…


Our Nature paper (rdcu.be/ei1NM) deep sequencing a 4-generation, 28-member family using multiple sequencing technologies to study transmission of all classes of genetic variation is out! Evan Eichler UW Genome Sciences HHMI


Congratulations to Glennis Logsdon (Penn Genetics/ Penn Epigenetics) on being named a 2025 Searle Scholar! tinyurl.com/ma4sp577